3MD9
 
 | | Structure of apo form of a periplasmic heme binding protein | | Descriptor: | BROMIDE ION, GLYCEROL, Hemin-binding periplasmic protein hmuT, ... | | Authors: | Mattle, D, Goetz, B.A, Locher, K.P. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
7RTH
 
 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNI
 
 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
6SNH
 
 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
4R9U
 
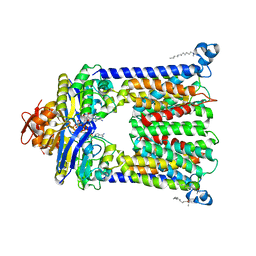 | | Structure of vitamin B12 transporter BtuCD in a nucleotide-bound outward facing state | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Veprintsev, D.B, Locher, K.P. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4ZRP
 
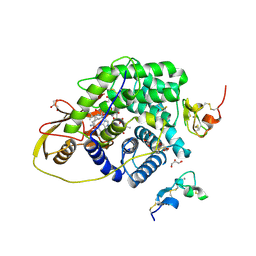 | | TC:CD320 | | Descriptor: | CALCIUM ION, CD320 antigen, CYANOCOBALAMIN, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2015-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of transcobalamin recognition by human CD320 receptor.
Nat Commun, 7, 2016
|
|
4ZRQ
 
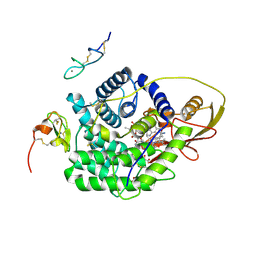 | |
5M3B
 
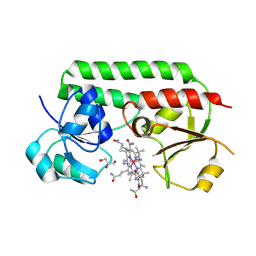 | | Structure of cobinamide-bound BtuF mutant W66L, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
5M34
 
 | | Structure of cobinamide-bound BtuF mutant W66Y, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
5M29
 
 | | Structure of cobinamide-bound BtuF, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
2ONS
 
 | | Crystal structure of A. fulgidus periplasmic binding protein ModA with bound tungstate | | Descriptor: | MAGNESIUM ION, NITRATE ION, TUNGSTATE(VI)ION, ... | | Authors: | Hollenstein, K, Frei, D.C, Locher, K.P. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of an ABC transporter in complex with its binding protein.
Nature, 446, 2007
|
|
2ONR
 
 | | Crystal structure of A. fulgidus periplasmic binding protein ModA with bound molybdate | | Descriptor: | MAGNESIUM ION, MOLYBDATE ION, NITRATE ION, ... | | Authors: | Hollenstein, K, Frei, D.C, Locher, K.P. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an ABC transporter in complex with its binding protein.
Nature, 446, 2007
|
|
2ONJ
 
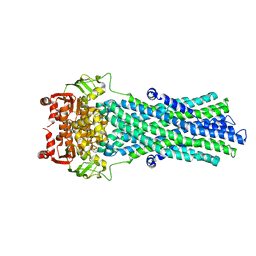 | |
2ONK
 
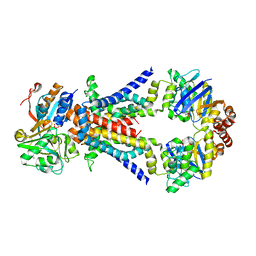 | | ABC transporter ModBC in complex with its binding protein ModA | | Descriptor: | MAGNESIUM ION, Molybdate/tungstate ABC transporter, ATP-binding protein, ... | | Authors: | Hollenstein, K, Frei, D.C, Locher, K.P. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of an ABC transporter in complex with its binding protein.
Nature, 446, 2007
|
|
2NQ2
 
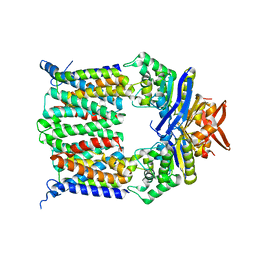 | | An inward-facing conformation of a putative metal-chelate type ABC transporter. | | Descriptor: | Hypothetical ABC transporter ATP-binding protein HI1470, Hypothetical ABC transporter permease protein HI1471 | | Authors: | Pinkett, H.P, Lee, A.T, Lum, P, Locher, K.P, Rees, D.C. | | Deposit date: | 2006-10-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An inward-facing conformation of a putative metal-chelate-type ABC transporter.
Science, 315, 2007
|
|
7A65
 
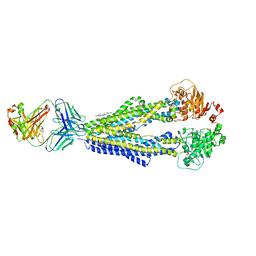 | |
7A6E
 
 | |
7A69
 
 | |
7A6F
 
 | |
7A6C
 
 | |
4G1U
 
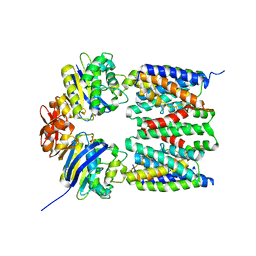 | | X-ray structure of the bacterial heme transporter HmuUV from Yersinia pestis | | Descriptor: | Hemin import ATP-binding protein HmuV, Hemin transport system permease protein hmuU, PHOSPHATE ION | | Authors: | Woo, J.-S, Goetz, B.A, Zeltina, A, Locher, K.P. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | X-ray structure of the Yersinia pestis heme transporter HmuUV.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FI3
 
 | | Structure of vitamin B12 transporter BtuCD-F in a nucleotide-bound state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Locher, K.P. | | Deposit date: | 2012-06-07 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.466 Å) | | Cite: | Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F.
Nature, 490, 2012
|
|
3NU1
 
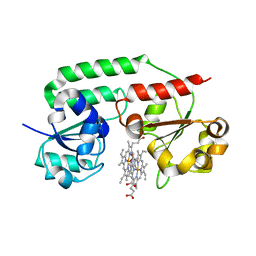 | | Structure of holo form of a periplasmic heme binding protein | | Descriptor: | Hemin-binding periplasmic protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mattle, D, Goetz, B.A, Woo, J.S, Locher, K.P. | | Deposit date: | 2010-07-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
3RCE
 
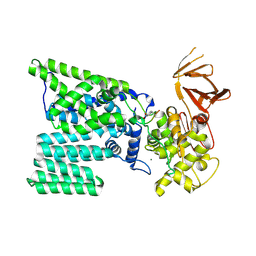 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
7OJH
 
 | | ABCG2 topotecan turnover-1 state | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, ... | | Authors: | Yu, Q, Ni, D, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | Deposit date: | 2021-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
