3I62
 
 | |
5IJX
 
 | |
3I5Y
 
 | |
5IHX
 
 | |
4OBC
 
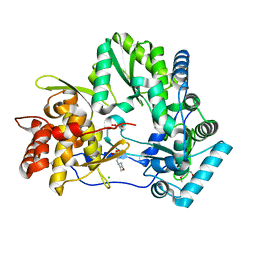 | | Crystal structure of HCV polymerase NS5b genotype 2a JFH-1 isolate with the S15G, C223H, V321I resistance mutations against the guanosine analog GS-0938 (PSI-3529238) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Abendroth, J, Appleby, T.C. | | Deposit date: | 2014-01-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
5XIM
 
 | |
5E0I
 
 | |
4E7A
 
 | |
4E76
 
 | |
4E78
 
 | |
8GIH
 
 | | Structure of Hepatitis B Virus Capsid Y132A mutant in complex with Compound 24 | | Descriptor: | (6S,8R)-N-(3-bromo-4-fluorophenyl)-8-fluoro-10-methyl-11-oxo-1,3,4,7,8,9,10,11-octahydro-2H-pyrido[4',3':3,4]pyrazolo[1,5-a][1,4]diazepine-2-carboxamide, CHLORIDE ION, Capsid protein | | Authors: | Shaffer, P.L. | | Deposit date: | 2023-03-14 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Diazepinone HBV capsid assembly modulators.
Bioorg Med Chem Lett, 72, 2022
|
|
9GIJ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound 5 | | Descriptor: | (2~{R})-3-(4-chlorophenyl)-2-[2-[(2~{R})-1-isoquinolin-4-ylcarbonylpyrrolidin-2-yl]ethanoyl-methyl-amino]-~{N}-methyl-propanamide, 3C-like proteinase nsp5 | | Authors: | Prasad, A, Schmitt, A, Preuss, F, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
9GIL
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
7XIM
 
 | |
1XIN
 
 | |
4XIM
 
 | |
5OLJ
 
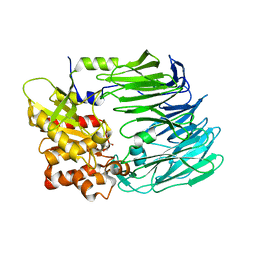 | | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 | | Descriptor: | Dipeptidyl peptidase IV, GLYCEROL | | Authors: | Fulop, V. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 and structure-activity relationships based on inhibitor profiling.
Eur J Med Chem, 139, 2017
|
|
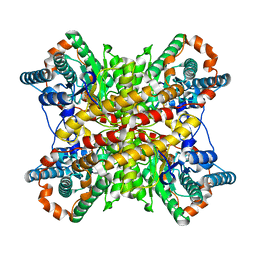
9XIM
 
 | |
3XIN
 
 | |
4DB2
 
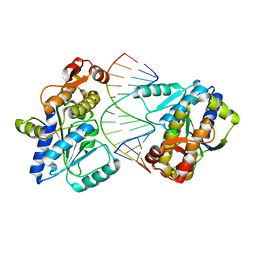 | | Mss116p DEAD-box helicase domain 2 bound to an RNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*GP*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
4DB4
 
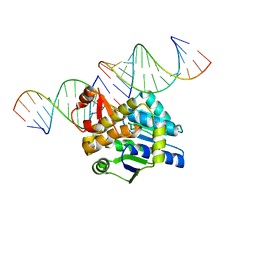 | | Mss116p DEAD-box helicase domain 2 bound to a chimaeric RNA-DNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
1XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
3XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, sorbitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
9C6M
 
 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase, K73M mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA ligase1 | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation
To Be Published
|
|
9C6L
 
 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, RNA ligase1, ... | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation
To Be Published
|
|
