6W4C
 
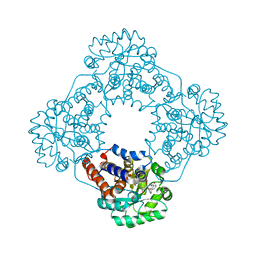 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
2YKG
 
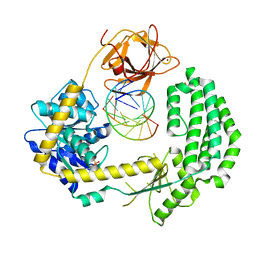 | | Structural insights into RNA recognition by RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
6HJS
 
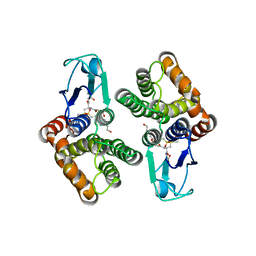 | | Crystal structure of glutathione transferase Omega 1C from Trametes versicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 2021
|
|
3KJL
 
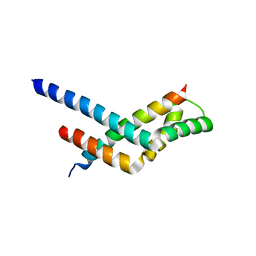 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1.
J.Biol.Chem., 285, 2010
|
|
3KIK
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
4AY2
 
 | | Capturing 5' tri-phosphorylated RNA duplex by RIG-I | | Descriptor: | 5'-R-PPP(GP*GP*CP*GP*CP*GP*GP*CP*UP*UP*CP*GP*GP*CP *CP*GP*CP*GP*CP*C)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-06-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor Rig-I.
Structure, 20, 2012
|
|
6HG7
 
 | | Crystal structure of a collagen II fragment containing the binding site of PEDF and COMP, (POG)4-LKG HRG FTG LQG-POG(4) | | Descriptor: | Collagen alpha-1(II) chain, SULFATE ION | | Authors: | Gebauer, J.M, Koehler, A, Dietmar, H, Gompert, M, Neundorf, I, Zaucke, F, Koch, M, Baumann, U. | | Deposit date: | 2018-08-22 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens.
Sci Rep, 8, 2018
|
|
6SR9
 
 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with oxyresveratrol | | Descriptor: | GLUTATHIONE, Uncharacterized protein, trans-oxyresveratrol | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SR8
 
 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SRB
 
 | | Crystal structure of glutathione transferase Omega 3C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SRA
 
 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with naringenin | | Descriptor: | GLUTATHIONE, R-naringenin, glutathione transferase | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6W35
 
 | |
4BPB
 
 | | STRUCTURAL INSIGHTS INTO RNA RECOGNITION BY RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
5EQE
 
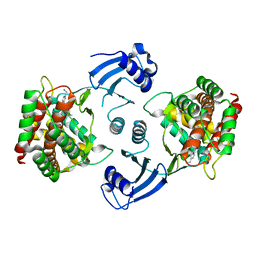 | | Crystal structure of choline kinase alpha-1 bound by [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine (compound 11) | | Descriptor: | Choline kinase alpha, [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5EQY
 
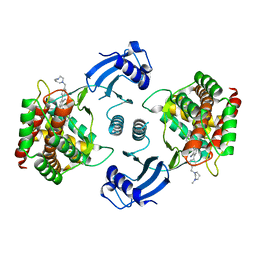 | | Crystal structure of choline kinase alpha-1 bound by 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile (compound 65) | | Descriptor: | 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5EQP
 
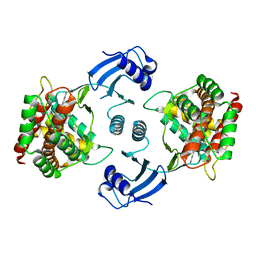 | | Crystal structure of choline kinase alpha-1 bound by 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline (compound 37) | | Descriptor: | 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5A10
 
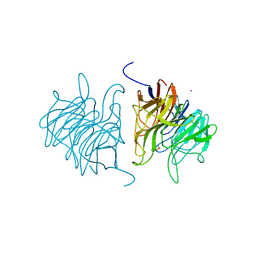 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group C2) | | Descriptor: | ACETATE ION, SODIUM ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
5A11
 
 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group P21) | | Descriptor: | IODIDE ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
3FWB
 
 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1 | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
3FWC
 
 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1, ... | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
3G8Q
 
 | |
