2ZCU
 
 | | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from escherichia coli | | Descriptor: | COPPER (II) ION, Uncharacterized oxidoreductase ytfG | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2ZCV
 
 | | Crystal structure of NADPH-dependent quinone oxidoreductase QOR2 complexed with NADPH from escherichia coli | | Descriptor: | COPPER (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2B8I
 
 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
7FEQ
 
 | | Cryo-EM structure of apo BsClpP at pH 6.5 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FES
 
 | | Cryo-EM structure of apo BsClpP at pH 4.2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FER
 
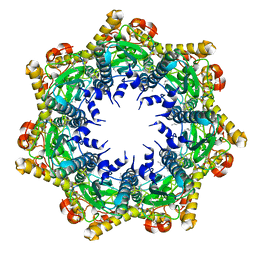 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 4.2 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FEP
 
 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 6.5 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
4FBB
 
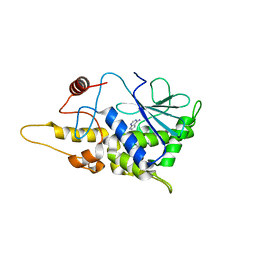 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBH
 
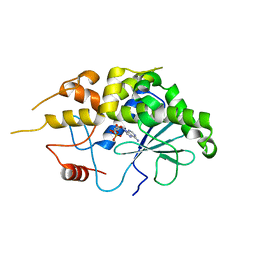 | | Structure of RIP from barley seeds | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBC
 
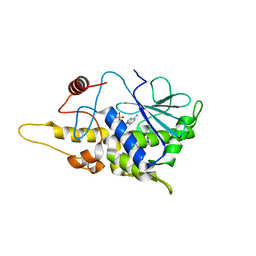 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FB9
 
 | | Structure of mutant RIP from barley seeds | | Descriptor: | Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBA
 
 | | Structure of mutant RIP from barley seeds in complex with adenine | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3G8E
 
 | | Crystal Structure of Rattus norvegicus Visfatin/PBEF/Nampt in Complex with an FK866-based inhibitor | | Descriptor: | 3-[(1E)-3-oxo-3-({4-[1-(phenylcarbonyl)piperidin-4-yl]butyl}amino)prop-1-en-1-yl]-1-beta-D-ribofuranosylpyridinium, Nicotinamide phosphoribosyltransferase | | Authors: | Kang, G.B, Bae, M.H, Kim, M.K, Im, I, Kim, Y.C, Eom, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Rattus norvegicus Visfatin/PBEF/Nampt in complex with an FK866-based inhibitor
Mol.Cells, 27, 2009
|
|
2PNV
 
 | | Crystal Structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SKCa) channel from Rattus norvegicus | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Kim, J.Y, Kim, M.K, Kang, G.B, Park, C.S, Eom, S.H. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SK(Ca)) channel from Rattus norvegicus.
Proteins, 70, 2008
|
|
3KU7
 
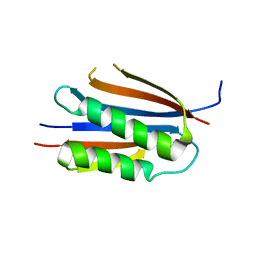 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Eom, S.H. | | Deposit date: | 2009-11-26 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
3MCD
 
 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Youn, H.S, Lee, J.G, An, J.Y, Jeon, H, Chun, J.S, Eom, S.H. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
3TT7
 
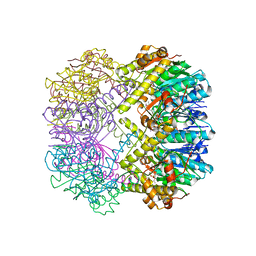 | |
3TT6
 
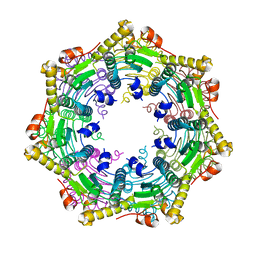 | |
4X3N
 
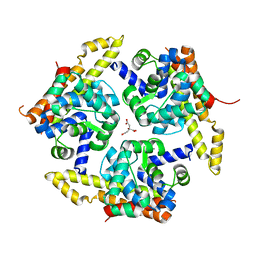 | | Crystal structure of 34 kDa F-actin bundling protein from Dictyostelium discoideum | | Descriptor: | CALCIUM ION, CITRIC ACID, Calcium-regulated actin-bundling protein | | Authors: | Kim, M.-K, Kim, J.-H, Kim, J.-S, Kang, S.-O. | | Deposit date: | 2014-12-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the 34 kDa F-actin-bundling protein ABP34 from Dictyostelium discoideum.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5GSC
 
 | |
5GZW
 
 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
4OOZ
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | Descriptor: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
