5WQ0
 
 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
5H3H
 
 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
6LVO
 
 | |
3AXX
 
 | |
7XRH
 
 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7YC0
 
 | | Acetylesterase (LgEstI) W.T. | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
6IYM
 
 | |
6JZL
 
 | |
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
6WMO
 
 | |
6WMN
 
 | |
6WMM
 
 | |
6OID
 
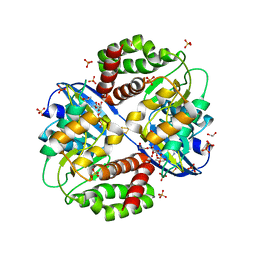 | | Redox Regulation of FN3K from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Wood, Z.A, Kadirvelraj, R, Shrestha, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily.
Sci.Signal., 13, 2020
|
|
2DS6
 
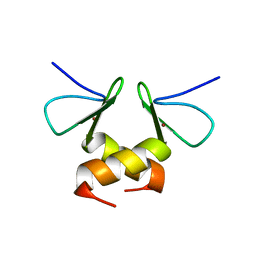 | | Structure of the ZBD in the tetragonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS7
 
 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
5XTF
 
 | |
5XTG
 
 | | Crystal structure of the cis-dihydrodiol naphthalene dehydrogenase NahB from Pseudomonas sp. MC1 in the presence of NAD+ and 2,3-dihydroxybiphenyl | | Descriptor: | 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase, BIPHENYL-2,3-DIOL, CITRIC ACID, ... | | Authors: | Park, A.K, Kim, H.-W. | | Deposit date: | 2017-06-19 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.318 Å) | | Cite: | Crystal structure of cis-dihydrodiol naphthalene dehydrogenase (NahB) from Pseudomonas sp. MC1: Insights into the early binding process of the substrate
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6AGQ
 
 | | Acetyl xylan esterase from Paenibacillus sp. R4 | | Descriptor: | ZINC ION, acetyl xylan esterase | | Authors: | Park, S, Lee, C.W, Lee, J.H. | | Deposit date: | 2018-08-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional characterization of a cold-active acetyl xylan esterase (PbAcE) from psychrophilic soil microbe Paenibacillus sp.
PLoS ONE, 13, 2018
|
|
7DVN
 
 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
1VWT
 
 | | T STATE HUMAN HEMOGLOBIN [ALPHA V96W], ALPHA AQUOMET, BETA DEOXY | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
1RVW
 
 | | R STATE HUMAN HEMOGLOBIN [ALPHA V96W], CARBONMONOXY | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PHOSPHATE ION, ... | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
5ZXE
 
 | | Structure of a consensus sequence derived from the FGF family | | Descriptor: | CHLORIDE ION, Consensus sequence based basic form of fibroblast growth factor, GLYCEROL, ... | | Authors: | Tripathi, S.K, Mandalaparthy, V, Ramaswamy, S, Gosavi, S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a consensus sequence derived from the FGF family
To be published
|
|
