1VDQ
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
1VDT
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
5CWV
 
 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
7YA8
 
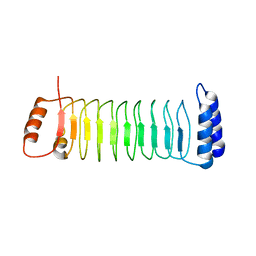 | | The crystal structure of IpaH2.5 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
7YA7
 
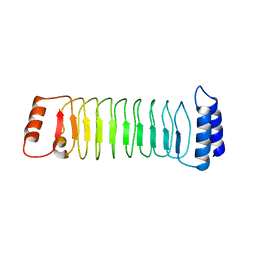 | | The crystal structure of IpaH1.4 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
7LO8
 
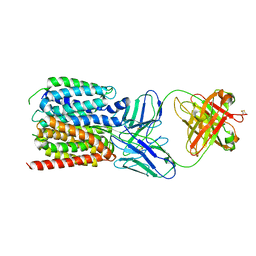 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO7
 
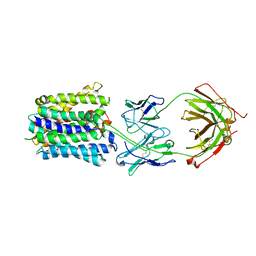 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
5DC9
 
 | | CRYSTAL STRUCTURE OF MONOBODY AS25/ABL1 SH2 DOMAIN COMPLEX, CRYSTAL B | | Descriptor: | AS25 monobody, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wojcik, J.B, Koide, A, Koide, S. | | Deposit date: | 2015-08-23 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface.
J.Biol.Chem., 291, 2016
|
|
5L4K
 
 | | The human 26S proteasome lid | | Descriptor: | 26S proteasome complex subunit DSS1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Schweitzer, A, Aufderheide, A, Rudack, T, Beck, F. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human 26S proteasome at a resolution of 3.9 angstrom.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8R8R
 
 | | Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc) | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*A)-3'), ... | | Authors: | Todesca, S, Sandmeir, F, Keidel, A, Conti, E. | | Deposit date: | 2023-11-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of human poly(A) polymerase recruitment by mPSF.
Rna, 30, 2024
|
|
2ZOE
 
 | | HA3 subcomponent of Clostridium botulinum type C progenitor toxin, complex with N-acetylneuramic acid | | Descriptor: | Hemagglutinin components HA3, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Kotani, M, Tonozuka, T, Ide, A, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-05-09 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
3AH4
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with galactose | | Descriptor: | Main hemagglutinin component, beta-D-galactopyranose | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH2
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH1
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylneuramic acid | | Descriptor: | Main hemagglutinin component, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
8EZG
 
 | | Monobody 12D1 bound to KRAS(G12D) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-10-31 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0M
 
 | | Monobody 12D5 bound to KRAS(G12D) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7QDS
 
 | | Apo human SKI complex in the open state | | Descriptor: | Helicase SKI2W, Tetratricopeptide repeat protein 37, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Bonneau, A, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
8TTE
 
 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTG
 
 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTF
 
 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTH
 
 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
7P0K
 
 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
7QE0
 
 | | 80S-bound human SKI complex in the open state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
