1A0A
 
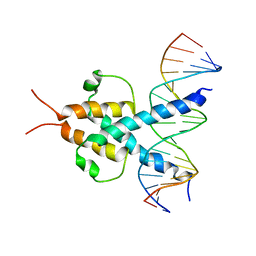 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
4ZKF
 
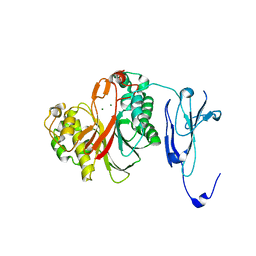 | | Crystal structure of human phosphodiesterase 12 | | Descriptor: | 2',5'-phosphodiesterase 12, MAGNESIUM ION | | Authors: | Kim, S.Y, Kohno, T, Mori, T, Kitano, K, Hakoshima, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human phosphodiesterase
To Be Published
|
|
6K3C
 
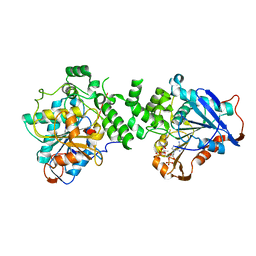 | | Crystal structure of class I PHA synthase (PhaC) mutant from Chromobacterium sp. USM2 bound to Coenzyme A. | | Descriptor: | COENZYME A, Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.074 Å) | | Cite: | Asymmetric Open-Closed Dimer Mechanism of Polyhydroxyalkanoate Synthase PhaC.
Iscience, 23, 2020
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
2F2U
 
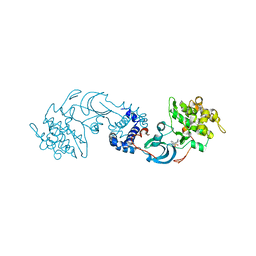 | | crystal structure of the Rho-kinase kinase domain | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Hakoshima, T. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil
Structure, 14, 2006
|
|
2H9V
 
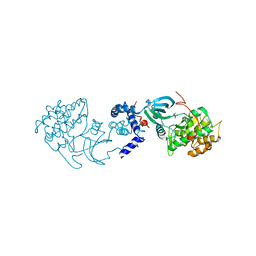 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1UIX
 
 | | Coiled-coil structure of the RhoA-binding domain in Rho-kinase | | Descriptor: | Rho-associated kinase | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase
J.Biol.Chem., 278, 2003
|
|
1DPF
 
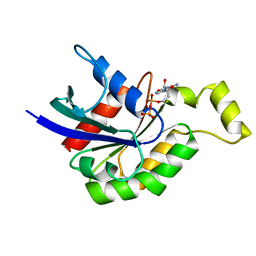 | | CRYSTAL STRUCTURE OF A MG-FREE FORM OF RHOA COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RHOA | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-12-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism.
J.Biol.Chem., 275, 2000
|
|
6KPD
 
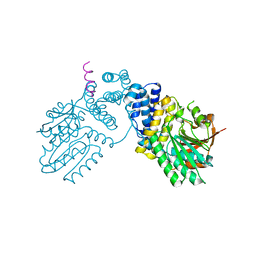 | |
6KPB
 
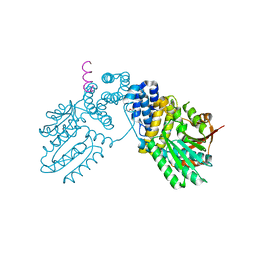 | |
5YIZ
 
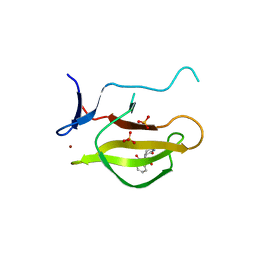 | |
5YJ0
 
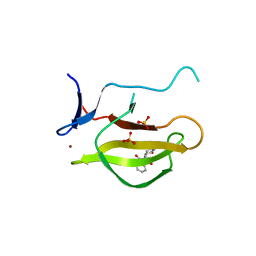 | |
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IS7
 
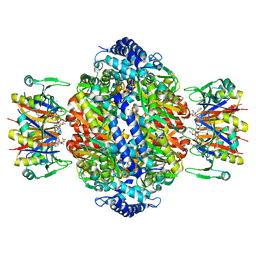 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5YJ1
 
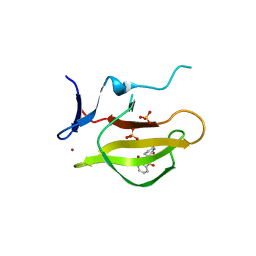 | | Mouse Cereblon thalidomide binding domain complexed with R-form thalidomide | | Descriptor: | 2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Mori, T, Hakoshima, T. | | Deposit date: | 2017-10-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of thalidomide enantiomer binding to cereblon
Sci Rep, 8, 2018
|
|
5B3H
 
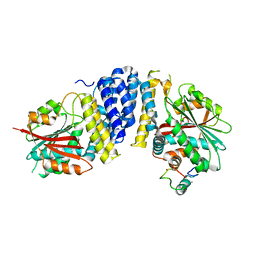 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | Descriptor: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | Authors: | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5B5V
 
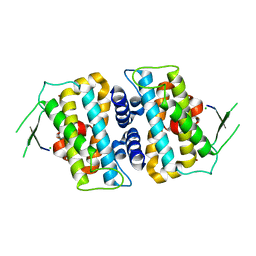 | | Structure of full-length MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B5W
 
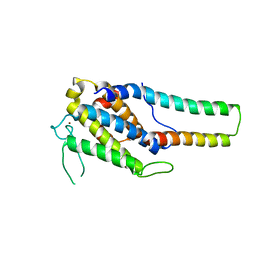 | | Crystal structure of MOB1-LATS1 NTR domain complex | | Descriptor: | MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B6B
 
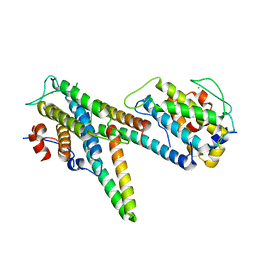 | | Complex of LATS1 and phosphomimetic MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ... | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.536 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B3G
 
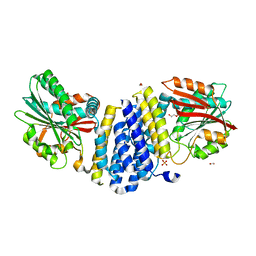 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Hirano, Y, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5XAV
 
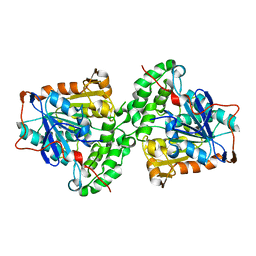 | | Structure of PhaC from Chromobacterium sp. USM2 | | Descriptor: | Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Arsad, H, Samian, M.R, Sudesh, K, Hakoshima, T. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics
Sci Rep, 7, 2017
|
|
2EMT
 
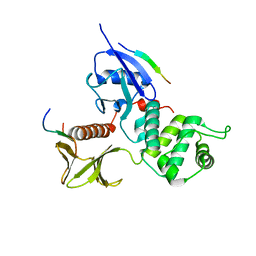 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | Descriptor: | P-selectin glycoprotein ligand 1, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
2EMS
 
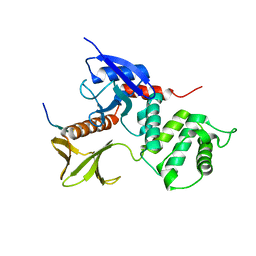 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule CD43 | | Descriptor: | Leukosialin, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins
J.Mol.Biol., 381, 2008
|
|
7DCH
 
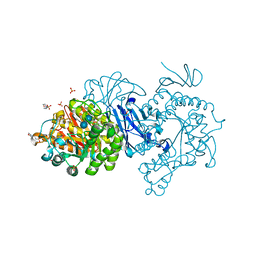 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with acarbose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glycosidase, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9C
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-13 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
