4D1Q
 
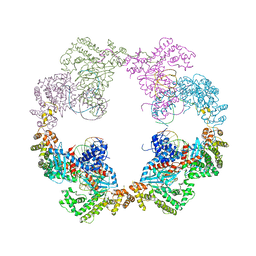 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
4ER8
 
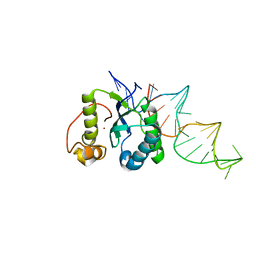 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
1AER
 
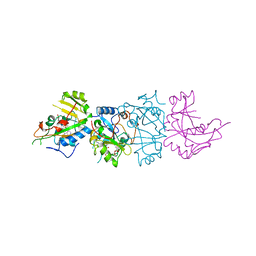 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH BETA-TAD | | Descriptor: | 2-(1,5-DIDEOXYRIBOSE)-4-AMIDO-THIAZOLE, ADENOSINE MONOPHOSPHATE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D.R. | | Deposit date: | 1995-12-11 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1BIS
 
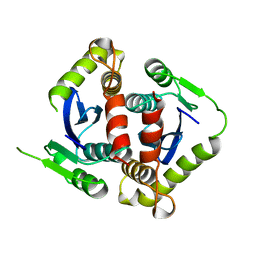 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIU
 
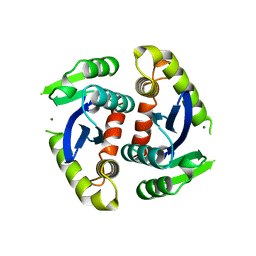 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
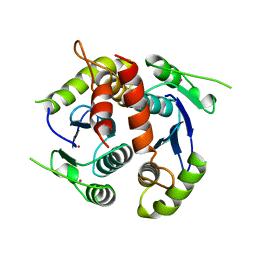 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6X68
 
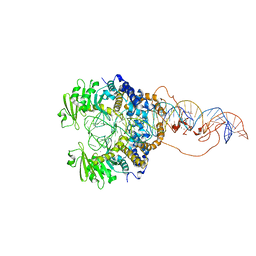 | | Cryo-EM structure of piggyBac transposase synaptic complex with hairpin DNA (SNHP) | | Descriptor: | CALCIUM ION, Transposase, ZINC ION, ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6XG8
 
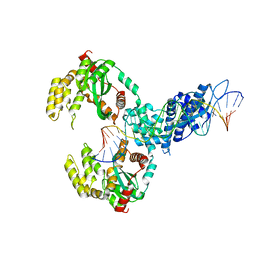 | | ISCth4 transposase, pre-cleaved complex, PCC | | Descriptor: | DNA (26-MER), Mutator family transposase | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
6XGW
 
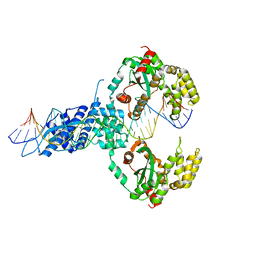 | | ISCth4 transposase, pre-reaction complex, PRC | | Descriptor: | DNA (32-MER), Mutator family transposase | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
6XGX
 
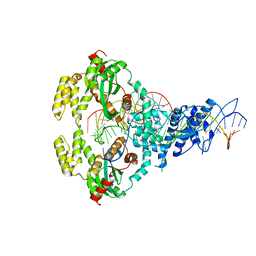 | | ISCth4 transposase, strand transfer complex 1, STC1 | | Descriptor: | DNA (21-MER), DNA (25-MER), DNA (47-MER), ... | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
6X67
 
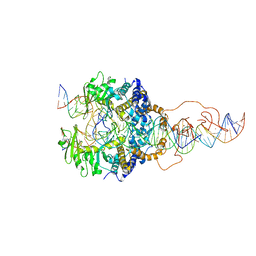 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | Descriptor: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
1YQV
 
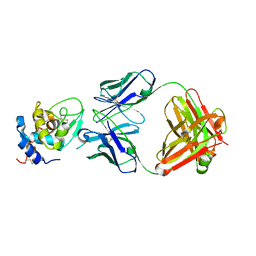 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2VJV
 
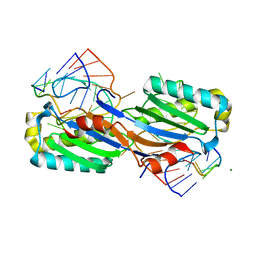 | | Crystal structure of the IS608 transposase in complex with left end 26-mer DNA hairpin and a 6-mer DNA representing the left end cleavage site | | Descriptor: | 5'-D(*DA*DA*DA*DG*DC*DC*DC*DC*DT*DA*DG*DC*DTP*DT *DT*DT*DA*DG*DC*DT*DA*DT*DG*DG*DG*DGP)-3', 5'-D(*DT*DA*DT*DT*DA*DCP)-3', MAGNESIUM ION, ... | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VJU
 
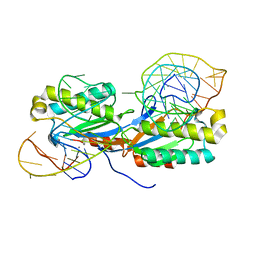 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2V0X
 
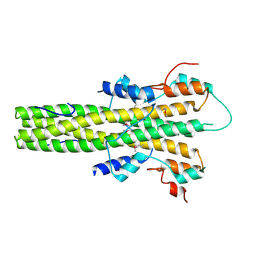 | | The dimerization domain of LAP2alpha | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | Authors: | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | Deposit date: | 2007-05-20 | | Release date: | 2007-06-26 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
2VIH
 
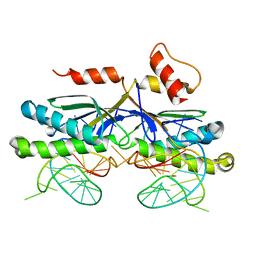 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
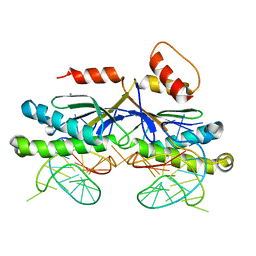 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VHG
 
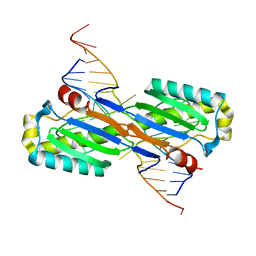 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | Descriptor: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
1AIH
 
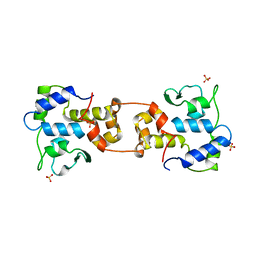 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1B6B
 
 | |
1IB1
 
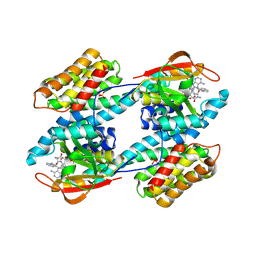 | | CRYSTAL STRUCTURE OF THE 14-3-3 ZETA:SEROTONIN N-ACETYLTRANSFERASE COMPLEX | | Descriptor: | 14-3-3 ZETA ISOFORM, COA-S-ACETYL TRYPTAMINE, SEROTONIN N-ACETYLTRANSFERASE | | Authors: | Obsil, T, Ghirlando, R, Klein, D.C, Ganguly, S, Dyda, F. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation.
Cell(Cambridge,Mass.), 105, 2001
|
|
1M55
 
 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | Descriptor: | CHLORIDE ION, Rep protein, ZINC ION | | Authors: | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
1MPE
 
 | |
1MVK
 
 | | X-ray structure of the tetrameric mutant of the B1 domain of streptococcal protein G | | Descriptor: | Immunoglobulin G binding protein G, SULFATE ION | | Authors: | Frank, M.K, Dyda, F, Dobrodumov, A, Gronenborn, A.M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Core mutations switch monomeric protein GB1 into an intertwined tetramer.
Nat.Struct.Biol., 9, 2002
|
|
