7RY6
 
 | | Solution NMR structural bundle of the first cyclization domain from yersiniabactin synthetase (Cy1) impacted by dynamics | | Descriptor: | HMWP2 nonribosomal peptide synthetase | | Authors: | Kancherla, A.K, Mishra, S.H, Marincin, K.A, Nerli, S, Sgourakis, N.G, Dowling, D.P, Bouvignies, G, Frueh, D.P. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global protein dynamics as communication sensors in peptide synthetase domains.
Sci Adv, 8, 2022
|
|
6U76
 
 | | Structure of methanesulfinate monooxygenase MsuC from Pseudomonas fluorescens. | | Descriptor: | methanesulfinate monooxygenase | | Authors: | Soule, J, Gnann, A.D, Parker, M.J, McKenna, K.C, Nguyen, S.V, Phan, N.T, Wicht, D.K, Dowling, D.P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | To be published
To Be Published
|
|
7M5W
 
 | | Crystal structure of the HMG-C1 domain of human capicua bound to DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*TP*TP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*AP*AP*AP*GP*C)-3'), ... | | Authors: | Webb, J.P, Liew, J.J.M, Gnann, A.D, Dowling, D.P. | | Deposit date: | 2021-03-25 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular basis of DNA recognition by the HMG-box-C1 module of Capicua
Biorxiv, 2022
|
|
6NHL
 
 | | Crystal structure of QueE from Escherichia coli | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Grell, T.A.J, Bell, B.N, Nguyen, C, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity.
Protein Sci., 28, 2019
|
|
7JTJ
 
 | |
7JUA
 
 | |
3RQD
 
 | | Ideal Thiolate-Zinc Coordination Geometry in Depsipeptide Binding to Histone Deacetylase 8 | | Descriptor: | Histone deacetylase 8, Largazole, POTASSIUM ION, ... | | Authors: | Cole, K.E, Dowling, D.P, Christianson, D.W. | | Deposit date: | 2011-04-28 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Structural basis of the antiproliferative activity of largazole, a depsipeptide inhibitor of the histone deacetylases.
J.Am.Chem.Soc., 133, 2011
|
|
6UUG
 
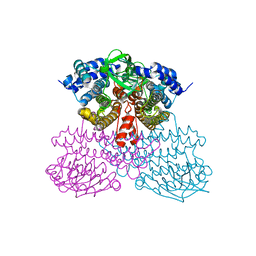 | | Structure of methanesulfinate monooxygenase MsuC from Pseudomonas fluorescens at 1.69 angstrom resolution | | Descriptor: | Putative dehydrogenase | | Authors: | Soule, J, Gnann, A.D, Gonzalez, R, Parker, M.J, McKenna, K.C, Nguyen, S.V, Phan, N.T, Wicht, D.K, Dowling, D.P. | | Deposit date: | 2019-10-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Structure and function of the two-component flavin-dependent methanesulfinate monooxygenase within bacterial sulfur assimilation.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
5TGS
 
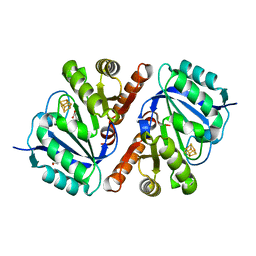 | | Crystal Structure of QueE from Bacillus subtilis with methionine bound | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
5TH5
 
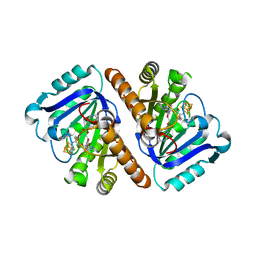 | | Crystal Structure of QueE from Bacillus subtilis with 6-carboxypterin-5'-deoxyadenosyl ester bound | | Descriptor: | 5'-O-(2-amino-4-oxo-1,4-dihydropteridine-6-carbonyl)adenosine, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
7JW9
 
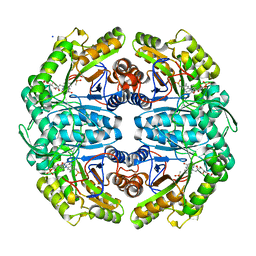 | | Ternary cocrystal structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, SODIUM ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JYB
 
 | | Binary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JV3
 
 | |
3GMZ
 
 | |
3GN0
 
 | |
3SJT
 
 | | Crystal structure of human arginase I in complex with the inhibitor Me-ABH, Resolution 1.60 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxyhexyl](trihydroxy)borate | | Authors: | Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
3SKK
 
 | | Crystal structure of human arginase I in complex with the inhibitor FABH, Resolution 1.70 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxy-6,6-difluorohexyl](trihydroxy)borate(1-) | | Authors: | Thorn, K.J, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
8SY8
 
 | | Crystal structure of TsaC | | Descriptor: | 4-formylbenzenesulfonate dehydrogenase TsaC | | Authors: | Boggs, D.G, Tian, J, Bridwell-Rabb, J. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The NADH recycling enzymes TsaC and TsaD regenerate reducing equivalents for Rieske oxygenase chemistry.
J.Biol.Chem., 299, 2023
|
|
