2KT2
 
 | | Structure of NmerA, the N-terminal HMA domain of Tn501 Mercuric Reductase | | Descriptor: | Mercuric reductase | | Authors: | Ledwidge, R, Danacea, F, Dotsch, V, Miller, S.M. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2KT3
 
 | | Structure of Hg-NmerA, Hg(II) complex of the N-terminal domain of Tn501 Mercuric Reductase | | Descriptor: | MERCURY (II) ION, Mercuric reductase | | Authors: | Miller, S.M, Ledwidge, R, Danacea, F, Dotsch, V. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2LIU
 
 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6H8C
 
 | | Structure of the human GABARAPL2 protein in complex with the UBA5 LIR motif | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 2, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Huber, J, Loehr, F, Gruber, J, Akutsu, M, Guentert, P, Doetsch, V, Rogov, V.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Autophagy, 16, 2020
|
|
3SJC
 
 | |
3SJA
 
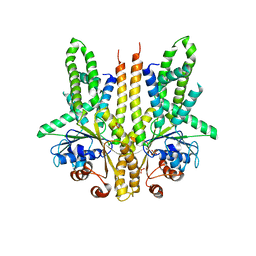 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJD
 
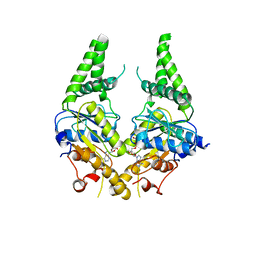 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6TBE
 
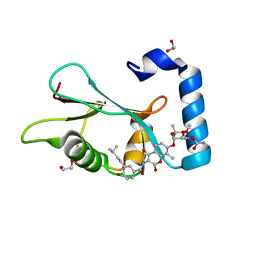 | | LC3A in complex with (3R,4S,5R,6R)-5-hydroxy-6-((4-hydroxy-3-(4-hydroxy-3-isopentylbenzamido)-8-methyl-2-oxo-2H-chromen-7-yl)oxy)-3-methoxy-2,2-dimethyltetrahydro-2H-pyran-4-yl carbamate | | Descriptor: | 1,2-ETHANEDIOL, Microtubule-associated proteins 1A/1B light chain 3A, NOVOBIOCIN | | Authors: | Kramer, J.S, Pogoryelov, D, Hartmann, M, Chaikuad, A, Proschak, E. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67008042 Å) | | Cite: | Demonstrating Ligandability of the LC3A and LC3B Adapter Interface.
J.Med.Chem., 64, 2021
|
|
4MRT
 
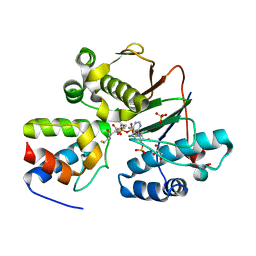 | | Structure of the Phosphopantetheine Transferase Sfp in Complex with Coenzyme A and a Peptidyl Carrier Protein | | Descriptor: | 4'-phosphopantetheinyl transferase sfp, COENZYME A, GLYCEROL, ... | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
4QVK
 
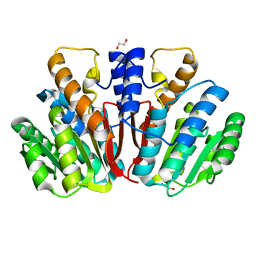 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | Descriptor: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
3SJB
 
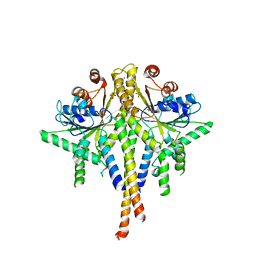 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
7OVC
 
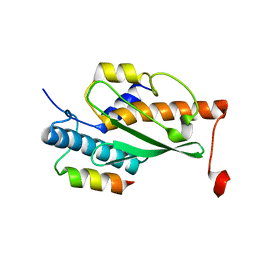 | | Structure of the human UFC1 protein in complex with the UBA5 C-terminal UFC1-binding motif. | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Wesch, W, Loehr, F, Rogova, N, Doetsch, V, Rogov, V.V. | | Deposit date: | 2021-06-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1.
Int J Mol Sci, 22, 2021
|
|
1GYF
 
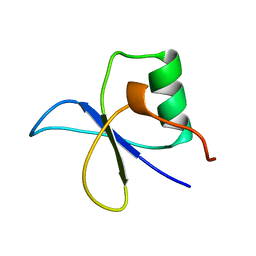 | | GYF DOMAIN FROM HUMAN CD2BP2 PROTEIN | | Descriptor: | PROTEIN (CYTOPLASMIC DOMAIN BINDING PROTEIN (CD2BP2)) | | Authors: | Freund, C, Doetsch, V, Nishizawa, K, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-30 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences.
Nat.Struct.Biol., 6, 1999
|
|
2RON
 
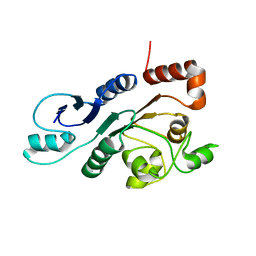 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
4XC2
 
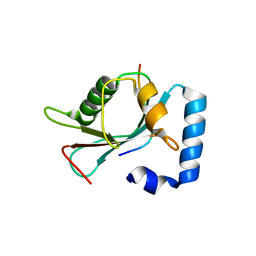 | | Crystal structure of GABARAP in complex with KBTBD6 LIR peptide | | Descriptor: | GABA(A) receptor-associated protein, Kelch repeat and BTB domain-containing protein 6 | | Authors: | Huber, J, Genau, H.M, Baschieri, F, Doetsch, V, Farhan, H, Rogov, V.V, Behrends, C, Akutsu, M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CUL3-KBTBD6/KBTBD7 Ubiquitin Ligase Cooperates with GABARAP Proteins to Spatially Restrict TIAM1-RAC1 Signaling.
Mol.Cell, 57, 2015
|
|
7SAF
 
 | |
7SAY
 
 | |
4YMG
 
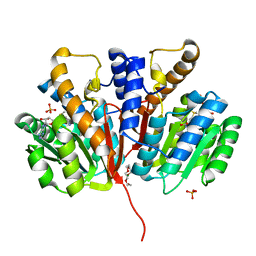 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4YMH
 
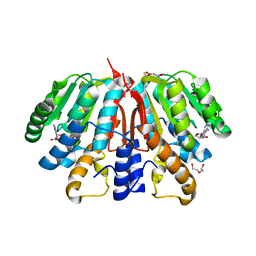 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
1A66
 
 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | Descriptor: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | Authors: | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | Deposit date: | 1998-03-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
6HB9
 
 | |
8P9C
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | 1,2-ETHANEDIOL, Darpin 1810 F11, Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
