6S8J
 
 | | Structure of ZEBOV GP in complex with 5T0180 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope Glycoprotein 1, Envelope glycoprotein, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S8I
 
 | | Structure of ZEBOV GP in complex with 3T0265 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Enveloped Glycoprotein 1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S8D
 
 | | Structure of ZEBOV GP in complex with 1T0227 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Heavy chain, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
4JDV
 
 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8P4T
 
 | | The spike complex of the Lujo Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Eilon-Ashkenazy, M, Diskin, R. | | Deposit date: | 2023-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The structure of the Lujo virus spike complex.
Nat Commun, 15, 2024
|
|
4ZJF
 
 | | Crystal structure of GP1 - the receptor binding domain of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein | | Authors: | Cohen-Dvashi, H, Cohen, N, Israeli, H, Diskin, R. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Molecular Mechanism for LAMP1 Recognition by Lassa Virus.
J.Virol., 89, 2015
|
|
6GC2
 
 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
6GJC
 
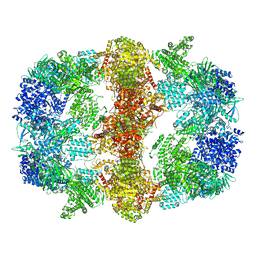 | | Structure of Mycobacterium tuberculosis Fatty Acid Synthase - I | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase | | Authors: | Elad, N, Baron, S, Shakked, Z, Zimhony, O, Diskin, R. | | Deposit date: | 2018-05-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution.
Nat Commun, 9, 2018
|
|
6GH8
 
 | | Crystal structure of GP1 domain of Lujo virus in complex with the first CUB domain of neuropilin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glycoprotein, ... | | Authors: | Cohen-Dvashi, H, Kilimnik, I, Diskin, R. | | Deposit date: | 2018-05-06 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for receptor recognition by Lujo virus.
Nat Microbiol, 3, 2018
|
|
5NSJ
 
 | | GP1 receptor-binding domain from Whitewater Arroyo mammarenavirus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex, ... | | Authors: | Shimon, A, Shani, O, Diskin, R. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural Basis for Receptor Selectivity by the Whitewater Arroyo Mammarenavirus.
J. Mol. Biol., 429, 2017
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
5OMI
 
 | | Crystal structure of GP2 from Lassa virus in a post fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Pre-glycoprotein polyprotein GP complex, ... | | Authors: | Shulman, A, Diskin, R. | | Deposit date: | 2017-07-31 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Variations in Core Packing of GP2 from Old World Mammarenaviruses in their Post-Fusion Conformations Affect Membrane-Fusion Efficiencies.
J.Mol.Biol., 431, 2019
|
|
6TM2
 
 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
7B0B
 
 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
7A5O
 
 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
4JDT
 
 | | Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Diskin, R, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OD6
 
 | | Crystal structure of p38alpha Y323T active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
2VSD
 
 | | crystal structure of CHIR-AB1 | | Descriptor: | CHIR AB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Arnon, T.I, Kaiser, J.T, Bjorkman, P.J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Chir-Ab1: A Primordial Avian Classical Fc Receptor.
J.Mol.Biol., 381, 2008
|
|
5T87
 
 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
5FT2
 
 | | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus- like particles at pH 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRE-GLYCOPROTEIN POLYPROTEIN GP COMPLEX | | Authors: | Li, S, Zhaoyang, S, Pryce, R, Parsy, M.L, Fehling, S.K, Schlie, K, Siebert, C.A, Garten, W, Bowden, T.A, Strecker, T, Huiskonen, J.T. | | Deposit date: | 2016-01-09 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Acidic Ph-Induced Conformations and Lamp1 Binding of the Lassa Virus Glycoprotein Spike.
Plos Pathog., 12, 2016
|
|
3OEF
 
 | | Crystal structure of Y323F inactive mutant of p38alpha MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
3ODY
 
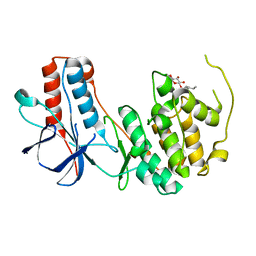 | | Crystal structure of p38alpha Y323Q active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
3ODZ
 
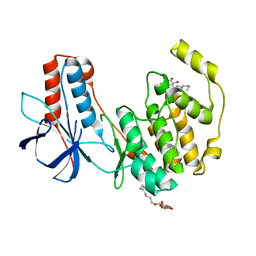 | | Crystal structure of P38alpha Y323R active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
6RBF
 
 | | Mucin 2 D3 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Intestinal Gel-Forming Mucins Polymerize by Disulfide-Mediated Dimerization of D3 Domains.
J.Mol.Biol., 431, 2019
|
|
6TM6
 
 | | MUC2 CysD1 domain | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-19 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
