1H8O
 
 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
4J2T
 
 | | Inhibitor-bound Ca2+ ATPase | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9bS)-6-(acetyloxy)-3a,4-bis(butanoyloxy)-3-hydroxy-3,6,9-trimethyl-8-{[(2E)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-7-yl octanoate, PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, ... | | Authors: | Paulsen, E.S, Villadsen, J, Tenori, E, Liu, H, Lie, M.A, Bonde, D.F, Bublitz, M, Olesen, C, Autzen, H.E, Dach, I, Sehgal, P, Moller, J.V, Schiott, B, Nissen, P, Christensen, S.B. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase.
J.Med.Chem., 56, 2013
|
|
2KJN
 
 | |
2LD0
 
 | |
2Q6P
 
 | | The Chemical Control of Protein Folding: Engineering a Superfolder Green Fluorescent Protein | | Descriptor: | Green fluorescent protein mutant 3 | | Authors: | Steiner, T, Hess, P, Bae, J.H, Wiltschi, B, Moroder, L, Budisa, N. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthetic biology of proteins: tuning GFPs folding and stability with fluoroproline.
Plos One, 3, 2008
|
|
2JQ1
 
 | | Phylloseptin-3 | | Descriptor: | Phylloseptin-3 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
2JX6
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin k by solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Mendonca Moraes, C, Verly, R.M, Resende, J.M, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Biophys.J., 96, 2009
|
|
2JPY
 
 | | Phylloseptin-2 | | Descriptor: | Phylloseptin-2 protein | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-24 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
2JQ0
 
 | | Phylloseptin-1 | | Descriptor: | Phylloseptin-1 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
2K9B
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Moraes, C.M, Verly, R.M, Resende, J.M, Aisenbrey, C, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy.
Biophys.J., 96, 2009
|
|
2LD2
 
 | |
2QOS
 
 | | Crystal structure of complement protein C8 in complex with a peptide containing the C8 binding site on C8 | | Descriptor: | Complement component 8, gamma polypeptide, Complement component C8 alpha chain | | Authors: | Lovelace, L.L, Chiswell, B, Slade, D.J, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of complement protein C8gamma in complex with a peptide containing the C8gamma binding site on C8alpha: Implications for C8gamma ligand binding.
Mol.Immunol., 45, 2008
|
|
2KJO
 
 | |
2W3L
 
 | | Crystal Structure of Chimaeric Bcl2-xL and Phenyl Tetrahydroisoquinoline Amide Complex | | Descriptor: | 1-(2-{[(3S)-3-(aminomethyl)-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}phenyl)-4-chloro-5-methyl-N,N-diphenyl-1H-pyrazole-3-carboxamide, APOPTOSIS REGULATOR BCL-2 | | Authors: | Porter, J, Payne, A, de Candole, B, Ford, D, Hutchinson, B, Trevitt, G, Turner, J, Edwards, C, Watkins, C, Whitcombe, I, Davis, J, Stubberfield, C, Fisher, M, Lamers, M. | | Deposit date: | 2008-11-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroisoquinoline Amide Substituted Phenyl Pyrazoles as Selective Bcl-2 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2YFY
 
 | | SERCA in the HnE2 State Complexed With Debutanoyl Thapsigargin | | Descriptor: | DEBUTANOYL THAPSIGARGIN, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sonntag, Y, Musgaard, M, Olesen, C, Schiott, B, Moller, J.V, Nissen, P, Thogersen, L. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutual Adaptation of a Membrane Protein and its Lipid Bilayer During Conformational Changes.
Nat.Commun., 2, 2011
|
|
2WLB
 
 | | Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria | | Descriptor: | ELECTRON TRANSFER PROTEIN 1, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Mueller, J.J, Hannemann, F, Schiffler, B, Bernhardt, R, Heinemann, U. | | Deposit date: | 2009-06-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Characterization of the Adrenodoxin-Like Domain of the Electron-Transfer Protein Etp1 from Schizosaccharomyces Pombe.
J.Inorg.Biochem., 105, 2011
|
|
7L23
 
 | |
3R3K
 
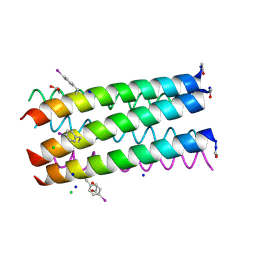 | | Crystal structure of a parallel 6-helix coiled coil | | Descriptor: | 1,2-ETHANEDIOL, CChex-Phi22 helix, CHLORIDE ION, ... | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.2009 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R46
 
 | | Crystal structure of a parallel 6-helix coiled coil CC-hex-D24 | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R4A
 
 | |
3R48
 
 | | Crystal structure of a hetero-hexamer coiled coil | | Descriptor: | GLYCEROL, coiled coil helix W22-L24H, coiled coil helix Y15-L24D | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0011 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R4H
 
 | |
3R47
 
 | | Crystal structure of a 6-helix coiled coil CC-hex-H24 | | Descriptor: | BROMIDE ION, coiled coil helix L24H | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5002 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
7NST
 
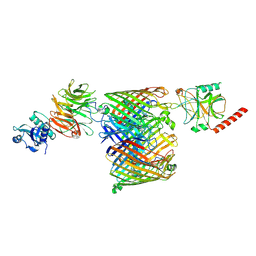 | | ColicinE9 partial translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7NSU
 
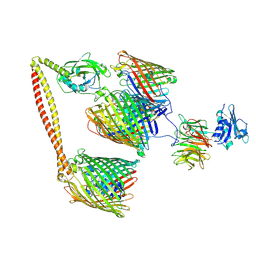 | | ColicinE9 intact translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB, ... | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
