4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5U
 
 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QGU
 
 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
3SR4
 
 | | Crystal Structure of Human DOT1L in Complex with a Selective Inhibitor | | Descriptor: | (2S)-2-azanyl-4-[[(2S,3S,4R,5R)-5-[6-(methylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Diao, J, Chen, P, Yao, Y, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-07-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
J.Am.Chem.Soc., 133, 2011
|
|
3M0D
 
 | | Crystal structure of the TRAF1:TRAF2:cIAP2 complex | | Descriptor: | Baculoviral IAP repeat-containing protein 3, TNF receptor-associated factor 1, TNF receptor-associated factor 2, ... | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-03-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation.
Mol.Cell, 38, 2010
|
|
3M06
 
 | | Crystal Structure of TRAF2 | | Descriptor: | TNF receptor-associated factor 2 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-03-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation.
Mol.Cell, 38, 2010
|
|
6NZV
 
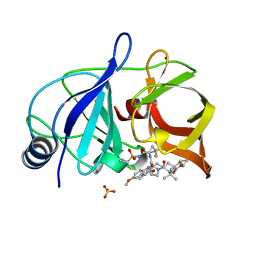 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3M0A
 
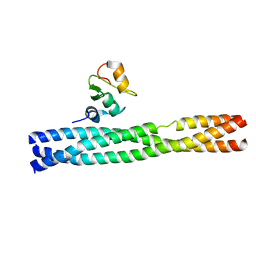 | | Crystal structure of TRAF2:cIAP2 complex | | Descriptor: | Baculoviral IAP repeat-containing protein 3, TNF receptor-associated factor 2, ZINC ION | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-03-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation.
Mol.Cell, 38, 2010
|
|
4NOK
 
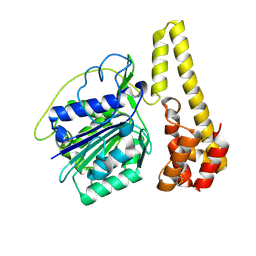 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOM
 
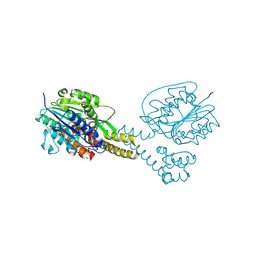 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOJ
 
 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
6INH
 
 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INI
 
 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6ING
 
 | |
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
5GZR
 
 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5H1Y
 
 | |
1RF3
 
 | | Structurally Distinct Recognition Motifs in Lymphotoxin-B Receptor and CD40 for TRAF-mediated Signaling | | Descriptor: | 24-residue peptide from Lymphotoxin-B Receptor, TNF receptor associated factor 3 | | Authors: | Li, C, Norris, P.S, Ni, C.Z, Havert, M.L, Chiong, E.M, Tran, B.R, Cabezas, E, Cheng, G, Reed, J.C, Satterthwait, A.C, Ware, C.F, Ely, K.R. | | Deposit date: | 2003-11-07 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling.
J.Biol.Chem., 278, 2003
|
|
8J1V
 
 | |
8J1T
 
 | |
6NZT
 
 | |
8FY7
 
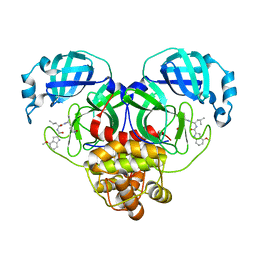 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FY6
 
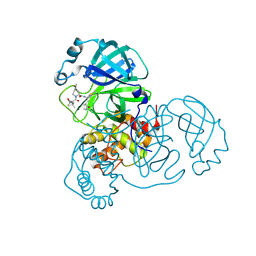 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | (1R,2S,5S)-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-N-{(2R)-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8H08
 
 | |
