3O7L
 
 | |
7QO4
 
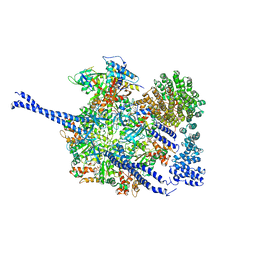 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
8HNA
 
 | |
8HN0
 
 | |
6L8O
 
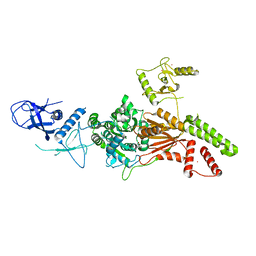 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
7JKS
 
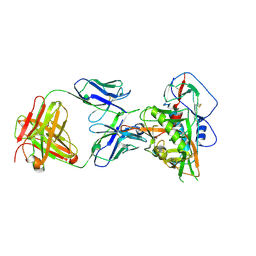 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
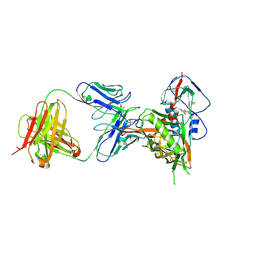 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
5TKK
 
 | |
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5V7J
 
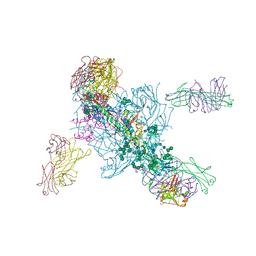 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
4LSJ
 
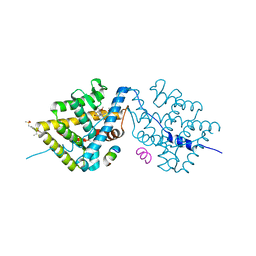 | | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Bound to a Dibenzoxapine Sulfonamide | | Descriptor: | D30 peptide, Glucocorticoid receptor, N-{3-[(1Z)-1-(10-methoxydibenzo[b,e]oxepin-11(6H)-ylidene)propyl]phenyl}methanesulfonamide | | Authors: | Carson, M, Luz, J.G, Clawson, D, Coghlan, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design.
J.Med.Chem., 57, 2014
|
|
4NRP
 
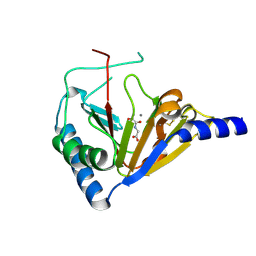 | |
4O7X
 
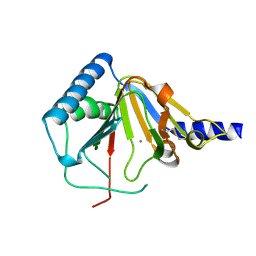 | |
4NRQ
 
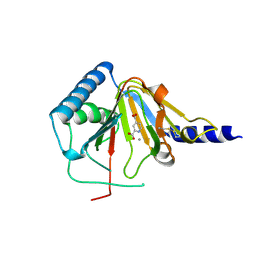 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRO
 
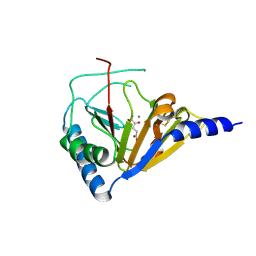 | |
4NRM
 
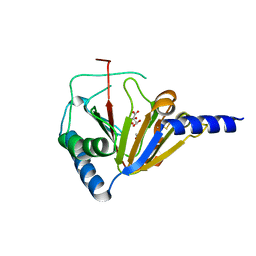 | |
6OSY
 
 | |
6OT1
 
 | |
1C5G
 
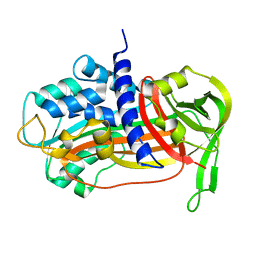 | | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Goldsmith, E.J. | | Deposit date: | 1999-12-07 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering of plasminogen activator inhibitor-1 to reduce the rate of latency transition.
Nat.Struct.Biol., 2, 1995
|
|
2BO3
 
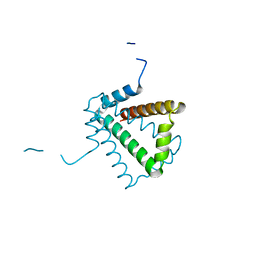 | | Crystal Structure of HP0242, a Hypothetical Protein from Helicobacter pylori | | Descriptor: | HYPOTHETICAL PROTEIN HP0242 | | Authors: | Sun, Y.-J, Tsai, J.-Y, Chen, B.-T. | | Deposit date: | 2005-04-07 | | Release date: | 2006-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Hp0242, a Hypothetical Protein from Helicobacter Pylori with a Novel Fold
Proteins: Struct., Funct., Bioinf., 62, 2006
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
9BLX
 
 | | Rhesus macaque ITS111.01 Fab in complex with SIV MPER peptide | | Descriptor: | Envelope glycoprotein gp160, GLYCEROL, ITS111.01 Heavy, ... | | Authors: | Gorman, J, Ahmadi, M, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Isolation and structure of broad SIV-neutralizing antibodies reveal a proximal helical MPER epitope recognized by a rhesus multi-donor class.
Cell Rep, 44, 2025
|
|
9BNS
 
 | |
9BP1
 
 | | Rhesus macaque ITS110.01 Fab in complex with SIV Env MPER peptide | | Descriptor: | Envelope glycoprotein gp160, ITS110.01 Heavy Chain, ITS110.01 Light Chain | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-05-06 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Isolation and structure of broad SIV-neutralizing antibodies reveal a proximal helical MPER epitope recognized by a rhesus multi-donor class.
Cell Rep, 44, 2025
|
|
