8TQV
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20403 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N-{[1-(methoxymethyl)cyclopropyl]methyl}-N-methylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TQG
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20419 | | Descriptor: | N-benzyl-2-{4-[4-(4,5-dimethoxy-1H-indole-2-carbonyl)piperazine-1-carbonyl]piperidin-1-yl}-6-methylpyrimidine-4-carboxamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TRY
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20348 | | Descriptor: | N-{(2S,3S)-4-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-3-hydroxy-1-phenylbutan-2-yl}-4-(2-methylbutan-2-yl)benzene-1-sulfonamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-10 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TR4
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20404 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N,N-dimethylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
5ZYA
 
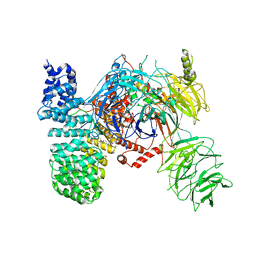 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
1SLN
 
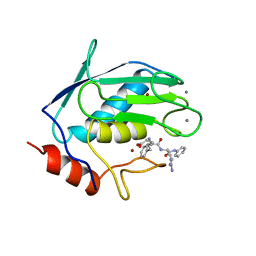 | |
1SLM
 
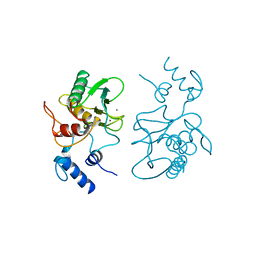 | |
1HFS
 
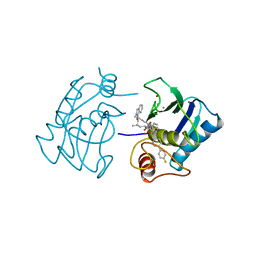 | |
4HFO
 
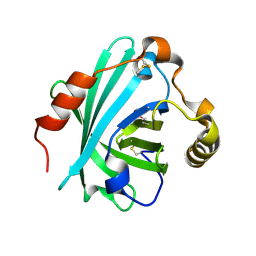 | | Biogenic amine-binding protein selenomethionine derivative | | Descriptor: | Biogenic amine-binding protein | | Authors: | Andersen, J.F, Xu, X, Chang, B, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QW4
 
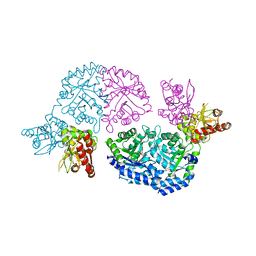 | | Structure of Leishmania donovani UMP synthase | | Descriptor: | CHLORIDE ION, UMP synthase, URIDINE-5'-MONOPHOSPHATE | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3QW3
 
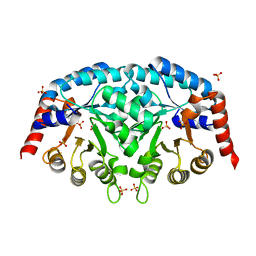 | | Structure of Leishmania donovani OMP decarboxylase | | Descriptor: | Orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, putative (Ompdcase-oprtase, putative), ... | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
8CPB
 
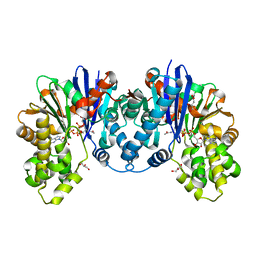 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
8CP9
 
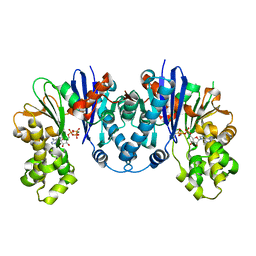 | |
8BRE
 
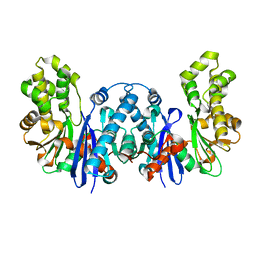 | |
8C0U
 
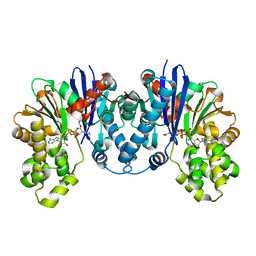 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with their natural substrates and products | | Descriptor: | (2~{R})-2-[(2~{S},3~{R},4~{R},5~{S},6~{R})-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
5J2N
 
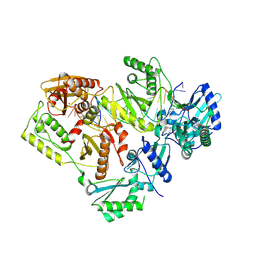 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated EFdA-MP at the P-(post-translocation) site and dTMP at the N-(pre-translocation) site | | Descriptor: | DNA (27-MER), DNA 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', MAGNESIUM ION, ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8RHI
 
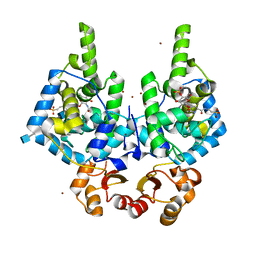 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa in a ternary complex bound to Bulgecin A and chito-tetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
8RHE
 
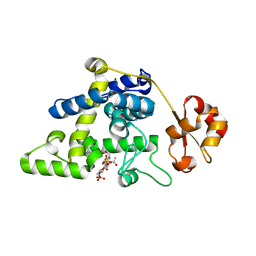 | |
8RHF
 
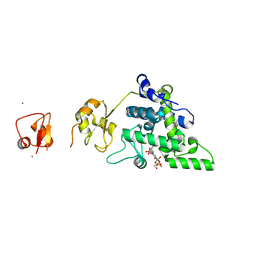 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa bound to the Natural Product Bulgecin A, with two LysM domains | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, DI(HYDROXYETHYL)ETHER, LysM peptidoglycan-binding domain-containing protein, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
1AO6
 
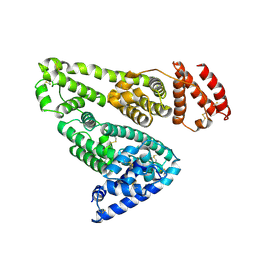 | |
1BM0
 
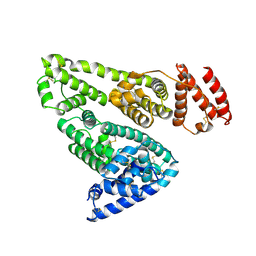 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
9FLK
 
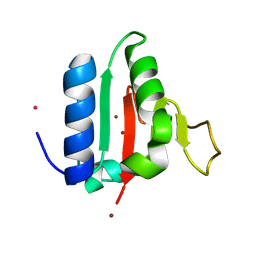 | | Crystal structure of the C-terminal domain of VldE from Streptococcus pneumoniae containing two zinc atoms at the binding site | | Descriptor: | CADMIUM ION, LysM domain-containing protein, ZINC ION | | Authors: | Acebron, I, Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of VldE (Spr1875), a Pneumococcal Two-State l,d-Endopeptidase with a Four-Zinc Cluster in the Active Site.
Acs Catalysis, 14, 2024
|
|
9FLH
 
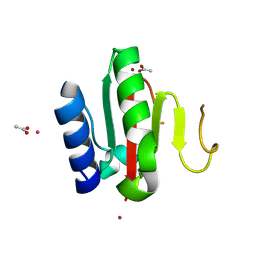 | | Crystal structure of the C-terminal domain of VldE from Streptococcus pneumoniae containing four zinc atoms at the binding site | | Descriptor: | ACETATE ION, CADMIUM ION, LysM domain-containing protein, ... | | Authors: | Acebron, I, Miguel-Ruano, V, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of VldE (Spr1875), a Pneumococcal Two-State l,d-Endopeptidase with a Four-Zinc Cluster in the Active Site.
Acs Catalysis, 14, 2024
|
|
9FLM
 
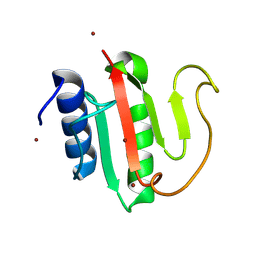 | | Crystal structure of the C-terminal domain of VldE from Streptococcus pneumoniae in a catalytically competent conformation | | Descriptor: | LysM domain-containing protein, ZINC ION | | Authors: | Miguel-Ruano, V, Acebron, I, P.de Jose, U, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of VldE (Spr1875), a Pneumococcal Two-State l,d-Endopeptidase with a Four-Zinc Cluster in the Active Site.
Acs Catalysis, 14, 2024
|
|
9FLL
 
 | | Crystal structure of the C-terminal domain of VldE from Streptococcus pneumoniae containing a zinc atom at the binding site | | Descriptor: | ACETATE ION, CADMIUM ION, LysM domain-containing protein, ... | | Authors: | Acebron, I, Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of VldE (Spr1875), a Pneumococcal Two-State l,d-Endopeptidase with a Four-Zinc Cluster in the Active Site.
Acs Catalysis, 14, 2024
|
|
