3SXO
 
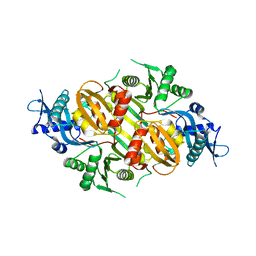 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7
To be Published
|
|
3RYO
 
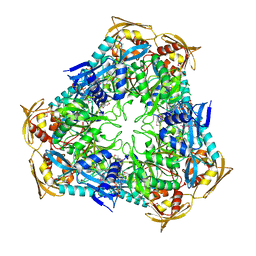 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
4KT6
 
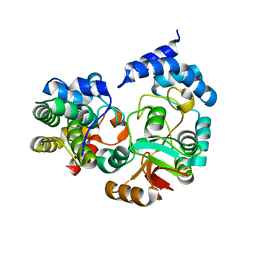 | | High-resolution crystal structure Streptococcus pyogenes beta-NAD+ glycohydrolase in complex with its endogenous inhibitor IFS reveals a water-rich interface | | Descriptor: | Nicotine adenine dinucleotide glycohydrolase, Putative uncharacterized protein | | Authors: | Yoon, J.Y, An, D.R, Yoon, H.-J, Kim, H.S, Lee, S.J, Im, H.N, Jang, J.Y, Suh, S.W. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution crystal structure of Streptococcus pyogenes beta-NAD(+) glycohydrolase in complex with its endogenous inhibitor IFS reveals a highly water-rich interface
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3QY8
 
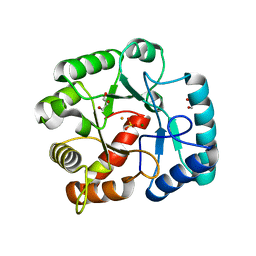 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY7
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY6
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, Tyrosine-protein phosphatase YwqE | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
4TX1
 
 | |
4QB9
 
 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5GTN
 
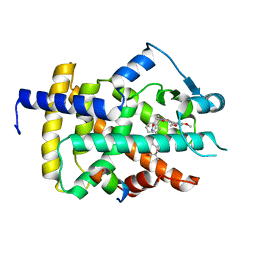 | | Human PPARgamma ligand binding dmain complexed with R35 | | Descriptor: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
5F8E
 
 | | Rv2258c-SAH | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8F
 
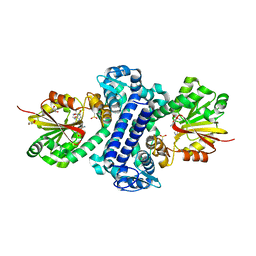 | | Rv2258c-SFG | | Descriptor: | GLYCEROL, Methyltransferase, SINEFUNGIN | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8C
 
 | | Rv2258c-unbound | | Descriptor: | GLYCEROL, Methyltransferase | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4Y4V
 
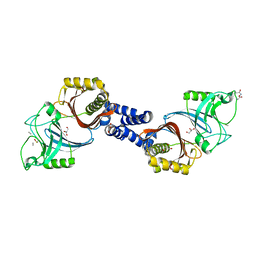 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3UY5
 
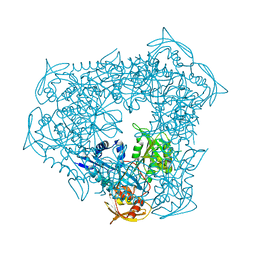 | | crystal structure of Eis from Mycobacterium tuberculosis | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, Suh, S.W. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
5GTO
 
 | | Human PPARgamma ligand binding dmain complexed with S35 | | Descriptor: | 2-[4-[5-[(1~{S})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
5GTP
 
 | |
3NIP
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase complexed with 1,6-diaminohexane | | Descriptor: | 3-guanidinopropionase, HEXANE-1,6-DIAMINE | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
3NIQ
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase | | Descriptor: | 3-guanidinopropionase, GLYCEROL, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
