1V57
 
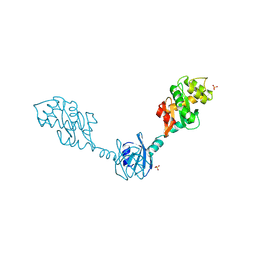 | | Crystal Structure of the Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4XVW
 
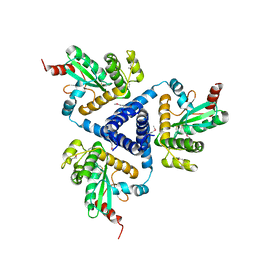 | |
4WF4
 
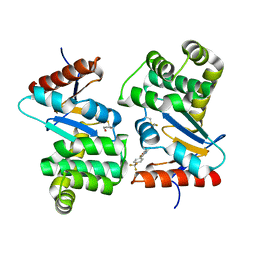 | | Crystal structure of E.Coli DsbA co-crystallised in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WEY
 
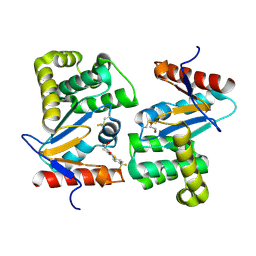 | | Crystal structure of E.Coli DsbA in complex with compound 17 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-serine, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WF5
 
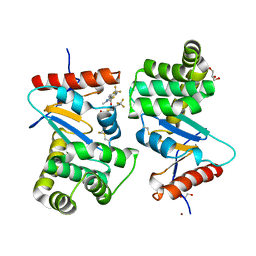 | | Crystal structure of E.Coli DsbA soaked with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, ... | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1WNH
 
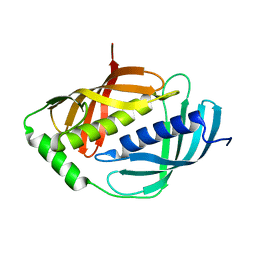 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
3F4R
 
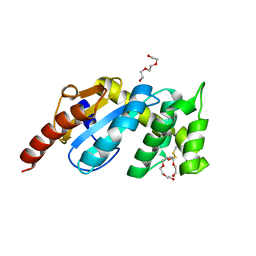 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis.
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
3F4S
 
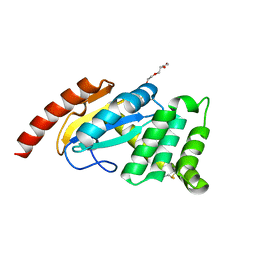 | |
3F4T
 
 | |
1UTE
 
 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, MU-OXO-DIIRON, ... | | Authors: | Guddat, L.W, Mcalpine, A, Hume, D, Hamilton, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1999-01-18 | | Release date: | 1999-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mammalian purple acid phosphatase.
Structure Fold.Des., 7, 1999
|
|
5VYO
 
 | |
3UX2
 
 | |
3UX3
 
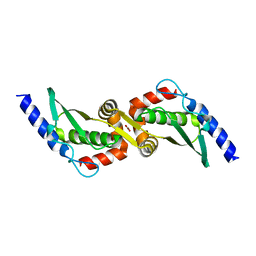 | | Crystal Structure of Domain-Swapped Fam96a minor dimer | | Descriptor: | ACETATE ION, MIP18 family protein FAM96A, ZINC ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1D4L
 
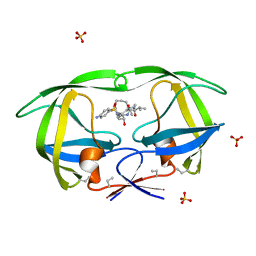 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1D4K
 
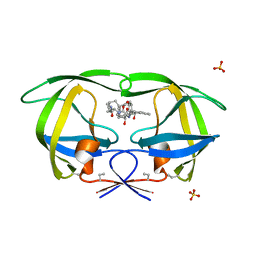 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1ACV
 
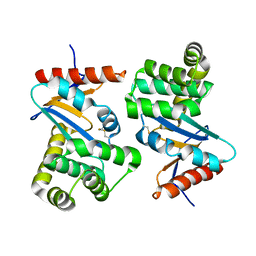 | | DSBA MUTANT H32S | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
1AC1
 
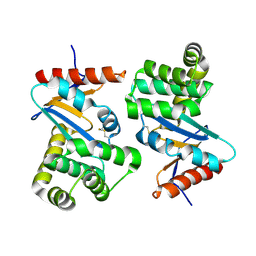 | | DSBA MUTANT H32L | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
3BCI
 
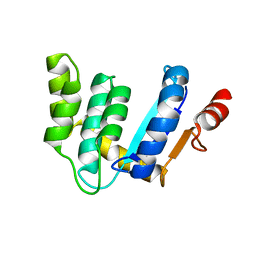 | |
3BXS
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxylic acid, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
1NUL
 
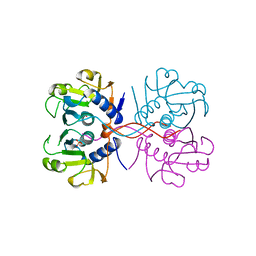 | | XPRTASE FROM E. COLI | | Descriptor: | MAGNESIUM ION, SULFATE ION, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1996-10-15 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli xanthine phosphoribosyltransferase.
Biochemistry, 36, 1997
|
|
2H0H
 
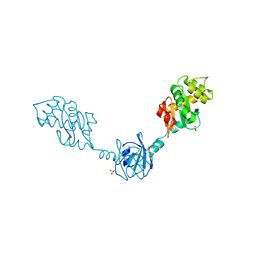 | | Crystal Structure of DsbG K113E mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0G
 
 | | Crystal Structure of DsbG T200M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
2H0I
 
 | | Crystal Structure of DsbG V216M mutant | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Hiniker, A, Heras, B, Martin, J.L, Stuckey, J, Bardwell, J.C.A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Short-circuiting divergent evolution: laboratory evolution of one disulfide isomerase to resemble another
To be Published
|
|
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
