7KQE
 
 | |
7KQB
 
 | | SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 5A6 heavy chain, Fab 5A6 light chain, ... | | Authors: | Asarnow, D, Charles, C, Cheng, Y. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
6LTW
 
 | | Crystal structure of Apo form of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8FE5
 
 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
1M64
 
 | | Crystal structure of Q363F mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mowat, C.G, Pankhurst, K.L, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-07-12 | | Release date: | 2002-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering water to act as an active site acid catalyst in a soluble fumarate reductase
Biochemistry, 41, 2002
|
|
6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
3B43
 
 | | I-band fragment I65-I70 from titin | | Descriptor: | Titin | | Authors: | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6TIJ
 
 | |
5K55
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
6TSG
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
5JYN
 
 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
2Z9T
 
 | | Crystal structure of the human beta-2 microglobulin mutant W60G | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Bolognesi, M, Bellotti, V, Corazza, A, Rennella, E, Gural, D, Mimmi, M.C, Betto, E, Pucillo, C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties
J.Mol.Biol., 378, 2008
|
|
6S9E
 
 | | Tubulin-GDP.AlF complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALUMINUM FLUORIDE, CALCIUM ION, ... | | Authors: | Oliva, M.A, Estevez-Gallego, J, Diaz, J.F, Prota, A.E, Steinmetz, M.O, Balaguer, F.A, Lucena-Agell, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural model for differential cap maturation at growing microtubule ends.
Elife, 9, 2020
|
|
6HMQ
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6HMB
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
1KRW
 
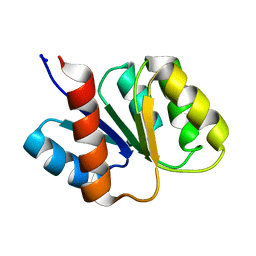 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
6EX1
 
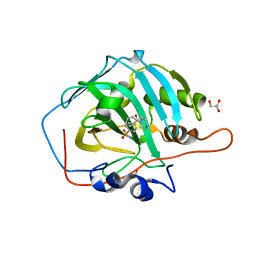 | | Crystal structure of human carbonic anhydrase I in complex with the 4-[(3S)-3 benzyl-4-(4-sulfamoylbenzoyl)piperazine -1-carbonyl]benzene-1-sulfonamide inhibitor | | Descriptor: | 4-[(3~{R})-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Ferraroni, M, Supuran, C.T, Chiapponi, D, Chiaramonte, N. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2-Benzylpiperazine: A new scaffold for potent human carbonic anhydrase inhibitors. Synthesis, enzyme inhibition, enantioselectivity, computational and crystallographic studies and in vivo activity for a new class of intraocular pressure lowering agents.
Eur J Med Chem, 151, 2018
|
|
6RNZ
 
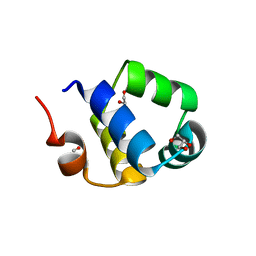 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RO6
 
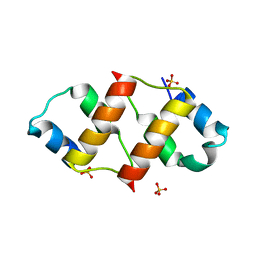 | | Crystal structure of the C-terminal dimerization domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC, SULFATE ION | | Authors: | Pignol, D, Arnoux, P, Siponen, M.I, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6FKM
 
 | |
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HV2
 
 | | MMP-13 in complex with the peptide IMISF | | Descriptor: | CALCIUM ION, Collagenase 3, GLYCEROL, ... | | Authors: | Mittl, P, Riedl, R, Hohl, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4B50
 
 | |
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
1LNB
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
