7DLD
 
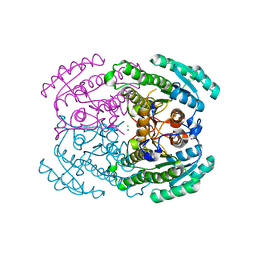 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | Descriptor: | Carbonyl Reductase, MAGNESIUM ION | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
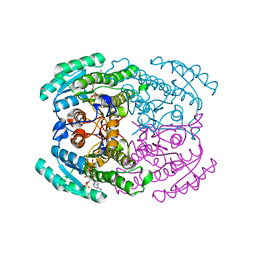 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
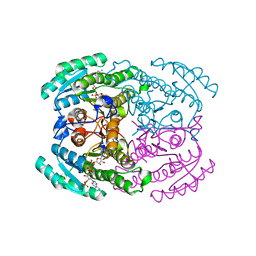 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7D6F
 
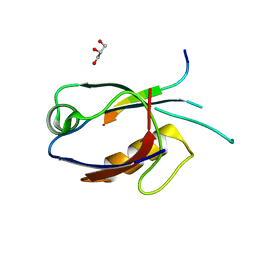 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
7CSP
 
 | | Structure of Ephexin4 IDPSH | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSR
 
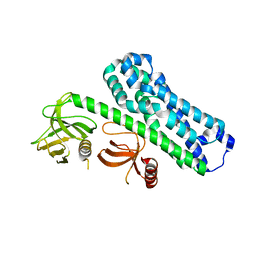 | | Structure of Ephexin4 R676L | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSO
 
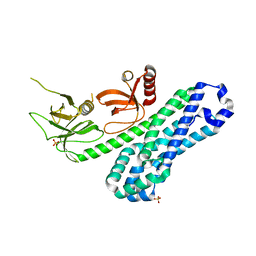 | | Structure of Ephexin4 DH-PH-SH3 | | Descriptor: | Rho guanine nucleotide exchange factor 16, SULFATE ION | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1XMJ
 
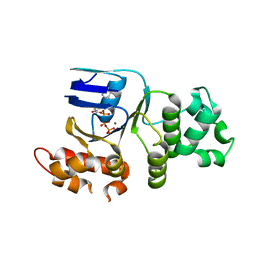 | | Crystal structure of human deltaF508 human NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
1XMI
 
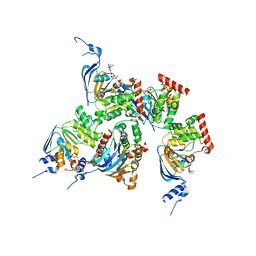 | | Crystal structure of human F508A NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
6TYY
 
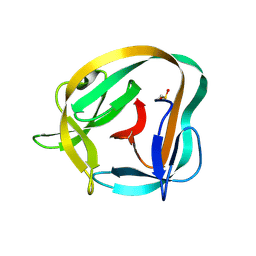 | | Hedgehog autoprocessing mutant D46H | | Descriptor: | Protein hedgehog | | Authors: | Li, H, Li, Z, Wang, C, Callahan, B.P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing.
J.Am.Chem.Soc., 141, 2019
|
|
6W8S
 
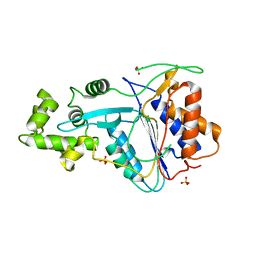 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8T
 
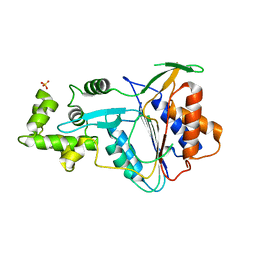 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8R
 
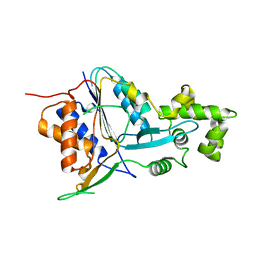 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6LPW
 
 | |
5WB6
 
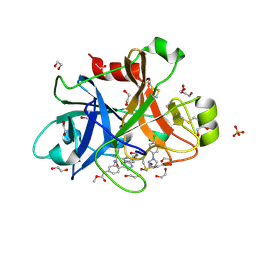 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4I5S
 
 | | Structure and function of sensor histidine kinase | | Descriptor: | Putative histidine kinase CovS; VicK-like protein | | Authors: | Cai, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains
Plos Biol., 11, 2013
|
|
4RBW
 
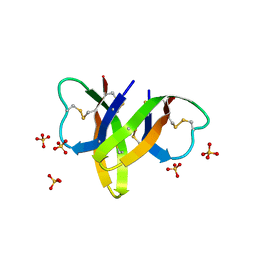 | | Crystal structure of human alpha-defensin 5, HD5 (Thr7Arg Glu21Arg mutant) | | Descriptor: | CHLORIDE ION, Defensin-5, SULFATE ION | | Authors: | Pazgier, M, Gohain, N, Tolbert, W.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a potent antibiotic peptide based on the active region of human defensin 5.
J.Med.Chem., 58, 2015
|
|
4RBX
 
 | |
7EQT
 
 | |
7EQS
 
 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
6LPV
 
 | |
9FI8
 
 | | SSU(head) structure derived from the SSU sample of the mitoribosome from T. gondii. | | Descriptor: | 30S ribosomal protein S5, putative, AP2 domain transcription factor AP2IX-6, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
