2G5C
 
 | | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, prephenate dehydrogenase | | Authors: | Sun, W, Singh, S, Zhang, R, Turnbull, J.L, Christendat, D. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus: Insights into the Catalytic Mechanism
J.Biol.Chem., 281, 2006
|
|
5HFE
 
 | |
4DX8
 
 | | ICAP1 in complex with KRIT1 N-terminus | | Descriptor: | BROMIDE ION, Integrin beta-1-binding protein 1, Krev interaction trapped protein 1 | | Authors: | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
1LQJ
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5HET
 
 | |
5HFD
 
 | |
5HFB
 
 | |
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
6JMT
 
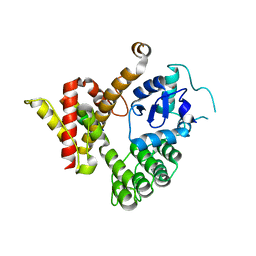 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
1MBE
 
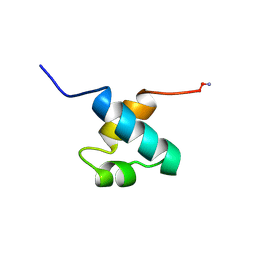 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
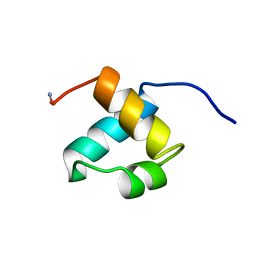 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
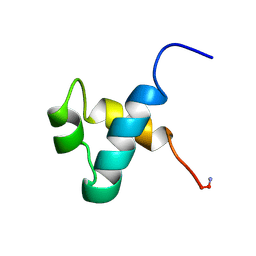 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
5IS2
 
 | |
1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
6JMU
 
 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
8JMY
 
 | |
1M1X
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHA VBETA3 BOUND TO MN2+ | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
8WB1
 
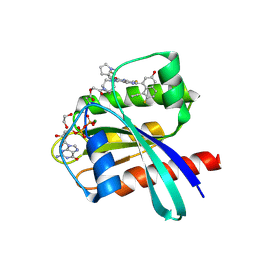 | | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},5~{R})-3-[7-(8-ethynyl-7-fluoranyl-3-oxidanyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]ethanone, GTPase KRas, ... | | Authors: | Zhou, L, Zhang, R, Shuai, T.B. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D)
To Be Published
|
|
5IT4
 
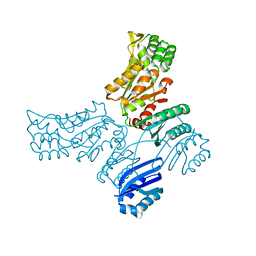 | |
1L5G
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN AVB3 IN COMPLEX WITH AN ARG-GLY-ASP LIGAND | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
4DXA
 
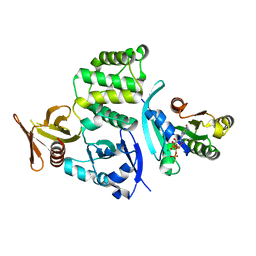 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
5JLR
 
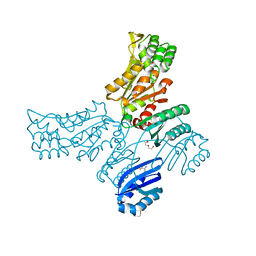 | | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, ... | | Authors: | Shree, S, Agrawal, A, Dubey, S, Ramachandran, R. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
To be published
|
|
