4MQF
 
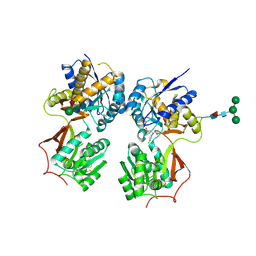 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist 2-hydroxysaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxysaclofen, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MQE
 
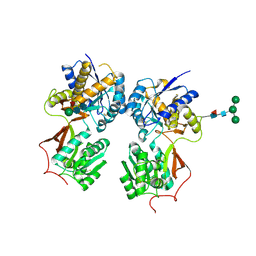 | | Crystal structure of the extracellular domain of human GABA(B) receptor in the apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4EC7
 
 | | Cobra NGF in complex with lipid | | Descriptor: | (2S)-1-hydroxy-3-(tetradecanoyloxy)propan-2-yl docosanoate, Venom nerve growth factor | | Authors: | Jiang, T, Wang, F, Tong, Q. | | Deposit date: | 2012-03-26 | | Release date: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into lipid-bound nerve growth factors
Faseb J., 26, 2012
|
|
4MRM
 
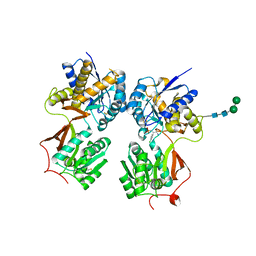 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist phaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
3SAY
 
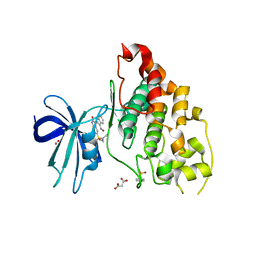 | | Crystal structure of human glycogen synthase kinase 3 beta (GSK3b) in complex with inhibitor 142 | | Descriptor: | (3Z)-N,N-diethyl-3-[(3E)-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-ylidene]-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide, (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, ... | | Authors: | Mazanetz, M.P, Cheng, R.K.Y, Rowan, F, Laughton, C.A, Barker, J.J, Fischer, P.M. | | Deposit date: | 2011-06-03 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Crystal structure of human glycogen synthase kinase 3 beta (GSK3b) in complex with inhibitor 142
To be Published
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEC
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UTN
 
 | | Crystal structure of Tum1 protein from Saccharomyces cerevisiae | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiosulfate sulfurtransferase TUM1 | | Authors: | Qiu, R, Wang, F, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tum1 protein from the yeast Saccharomyces cerevisiae.
Protein Pept.Lett., 19, 2012
|
|
3UEE
 
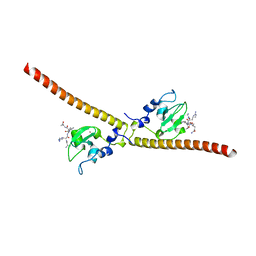 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UED
 
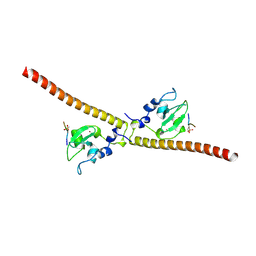 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEF
 
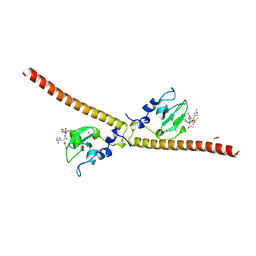 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
2L3N
 
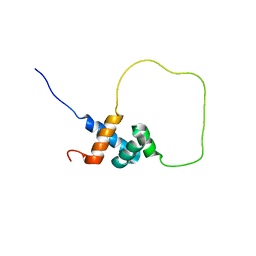 | | Solution structure of Rap1-Taz1 fusion protein | | Descriptor: | DNA-binding protein rap1,Telomere length regulator taz1 | | Authors: | Zhou, Z.R, Wang, F, Chen, Y, Lei, M, Hu, H. | | Deposit date: | 2010-09-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7DC1
 
 | |
5U36
 
 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
7JR9
 
 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JRJ
 
 | | Chlamydomonas reinhardtii radial spoke head and neck (recombinant) | | Descriptor: | Flagellar radial spoke protein 1, Flagellar radial spoke protein 10, Flagellar radial spoke protein 2, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Zhang, N, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L5W
 
 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5X
 
 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7Y40
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7YAO
 
 | |
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7XQK
 
 | | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, G1/S-specific cyclin-E1, GLYCEROL, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus.
To Be Published
|
|
