8SIR
 
 | |
8SIS
 
 | |
8SIT
 
 | |
8SIQ
 
 | |
2DV9
 
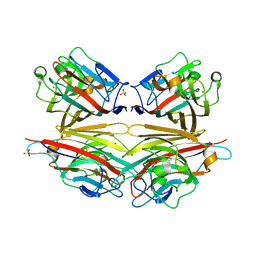 | | Crystal structure of peanut lectin GAL-BETA-1,3-GAL complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVG
 
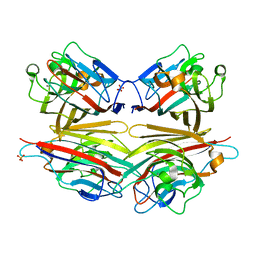 | | Crystal structure of peanut lectin GAL-ALPHA-1,6-GLC complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVA
 
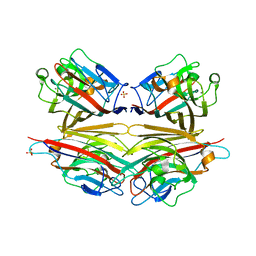 | | Crystal structure of peanut lectin GAL-BETA-1,3-GALNAC-ALPHA-O-ME (Methyl-T-antigen) complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
7PQE
 
 | |
2JWZ
 
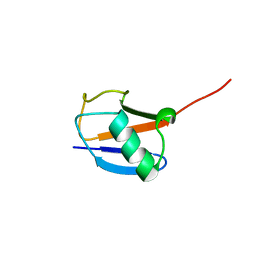 | | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins | | Descriptor: | Ubiquitin | | Authors: | Haririnia, A, Verma, R, Purohit, N, Twarog, M, Deshaies, R, Bolon, D, Fushman, D. | | Deposit date: | 2007-10-31 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins.
J.Mol.Biol., 375, 2008
|
|
7LX2
 
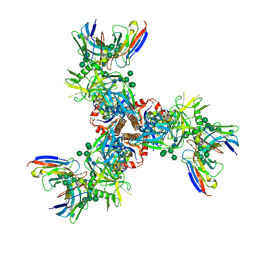 | | Cryo-EM structure of ConSOSL.UFO.664 (ConS) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LX3
 
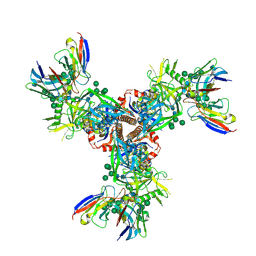 | | Cryo-EM structure of EDC-crosslinked ConSOSL.UFO.664 (ConS-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXM
 
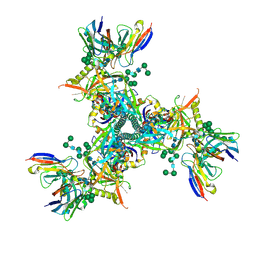 | | Cryo-EM structure of ConM SOSIP.v7 (ConM) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXN
 
 | | Cryo-EM structure of EDC-crosslinked ConM SOSIP.v7 (ConM-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
2LAH
 
 | | Solution NMR Structure of Mitotic checkpoint serine/threonine-protein kinase BUB1 N-terminal domain from Homo sapiens, Northeast Structural Genomics Consortium Target HR5460A (Methods Development) | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 | | Authors: | Liu, G, Xiao, R, Lee, H, Hamilton, K, Acton, T.B, Ciccosanti, C, Everett, J.K, Shastry, R.T, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium, n, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR5460A
To be Published
|
|
5LIO
 
 | |
4A7Q
 
 | | Structure of human I113T SOD1 mutant complexed with 4-(4-methyl-1,4- diazepan-1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-Ray Crystallography and Computational Docking for the Detection and Development of Protein-Ligand Interactions.
Curr.Med.Chem., 20, 2013
|
|
8SDH
 
 | |
8SDG
 
 | |
8SDF
 
 | |
4N7X
 
 | |
7ZP7
 
 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
7ZP6
 
 | |
7ZP5
 
 | |
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
8DGX
 
 | |
