6XV8
 
 | |
6XVI
 
 | |
7ES2
 
 | | a mutant of glycosyktransferase in complex with UDP and Reb D | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, rebaudioside D | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERX
 
 | | Glycosyltransferase in complex with UDP and STB | | Descriptor: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES1
 
 | | glycosyltransferase in complex with UDP and ST | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, steviol-19-o-glucoside | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES0
 
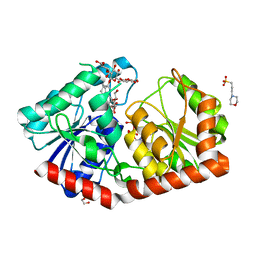 | | a rice glycosyltransferase in complex with UDP and REX | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERY
 
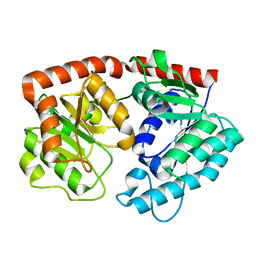 | | apo form of the glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
5OOO
 
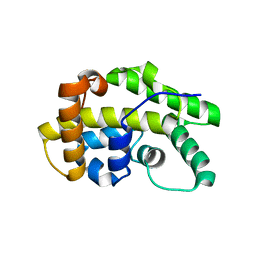 | |
5OO4
 
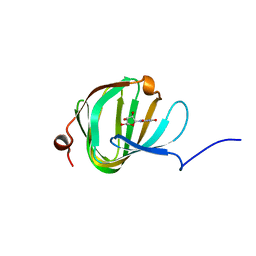 | | Streptomyces PAC13 with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OO5
 
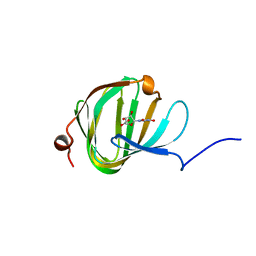 | | Streptomyces PAC13 with uridine uronic acid | | Descriptor: | Putative cupin_2 domain-containing isomerase, uridine uronic acid | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OOA
 
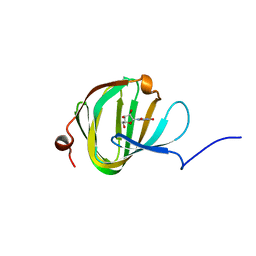 | | Streptomyces PAC13 (Y89F) with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O77
 
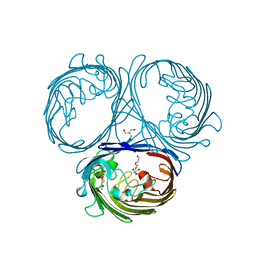 | | Klebsiella pneumoniae OmpK35 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | Authors: | van den berg, B, Pathania, M, Zahn, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5OO8
 
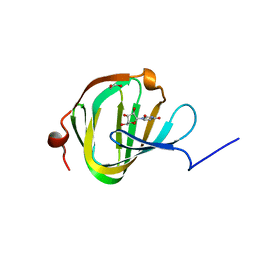 | | Streptomyces PAC13 (H42Q) with uridine | | Descriptor: | 1,2-ETHANEDIOL, Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O79
 
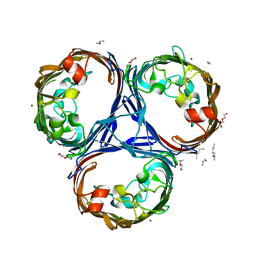 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5OO9
 
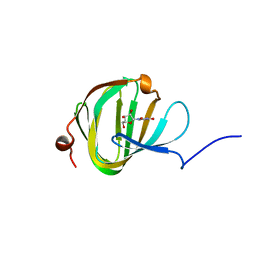 | | Streptomyces PAC13 (Y55F) with uridine | | Descriptor: | Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4B9E
 
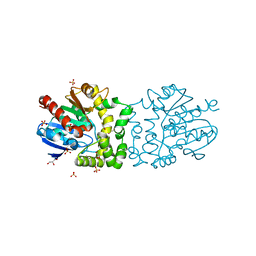 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B9A
 
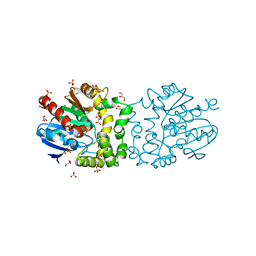 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1ZEN
 
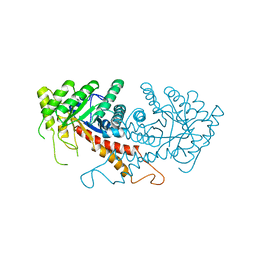 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Descriptor: | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE, ZINC ION | | Authors: | Cooper, S.J, Leonard, G.A, Hunter, W.N. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
Structure, 4, 1996
|
|
6ENE
 
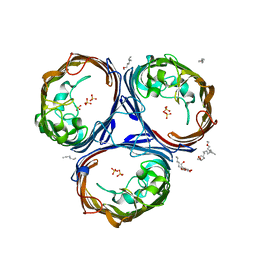 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
6QFA
 
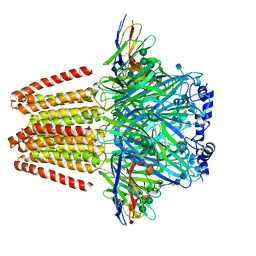 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZDH
 
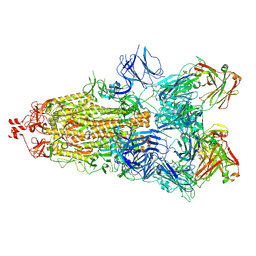 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Z1X
 
 | |
