9E9I
 
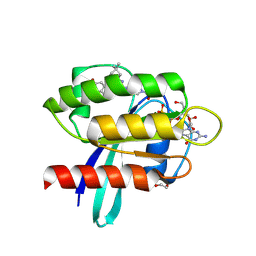 | | Crystal Structure of human KRAS G12C covalently bound to nopinone-derived naphthol compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(4P,6R,8R)-3-fluoro-4-(3-hydroxynaphthalen-1-yl)-7,7-dimethyl-5,6,7,8-tetrahydro-6,8-methanoquinolin-2-yl]-2,6-diazaspiro[3.4]octan-2-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2024-11-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Identification of Structurally Novel KRAS G12C Inhibitors through Covalent DNA-Encoded Library Screening.
J.Med.Chem., 68, 2025
|
|
9JK4
 
 | | putative E2P of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-15 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JK3
 
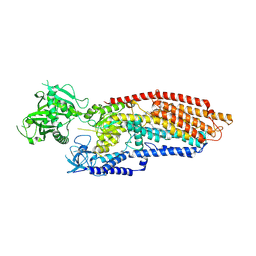 | | putative MgE2P of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJN
 
 | | putative MgE1-ATP of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJO
 
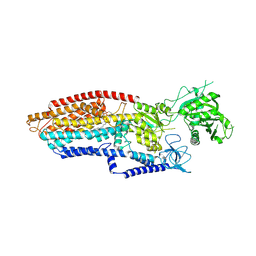 | | putative MgE1P-ADP of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JK5
 
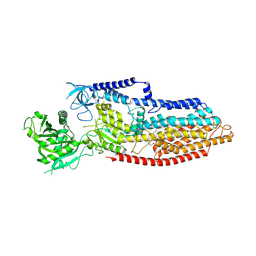 | | putative MgE1-ATP-H of TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-15 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
9JJK
 
 | | putative MgE1 of human TMEM94 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum magnesium-transporting P-type ATPase | | Authors: | Gong, D.S. | | Deposit date: | 2024-09-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural analysis of full-length human transmembrane protein 94 argues against its classification as a P-type Mg 2+ ATPase.
Cell Discov, 11, 2025
|
|
6ZDJ
 
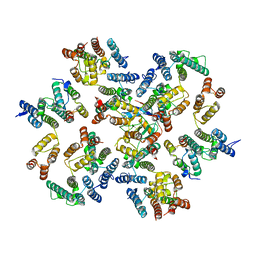 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8HVN
 
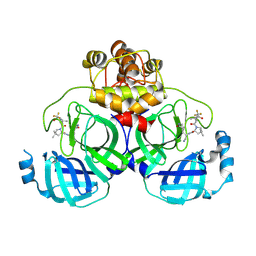 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVL
 
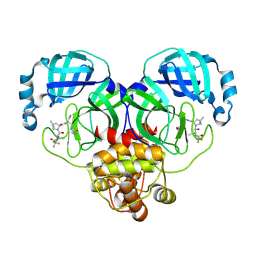 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVO
 
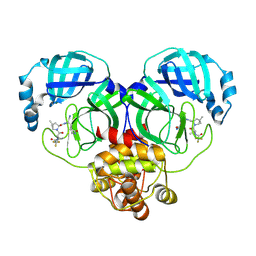 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVM
 
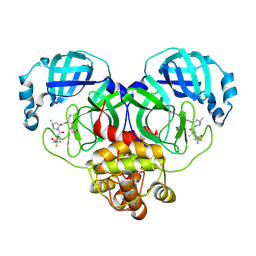 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVK
 
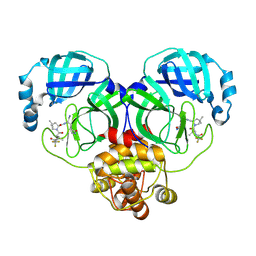 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HZR
 
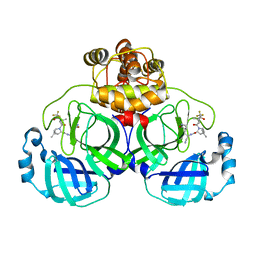 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
6M31
 
 | |
4NQJ
 
 | | Structure of coiled-coil domain | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, E3 ubiquitin-protein ligase TRIM69 | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural insights into the TRIM family of ubiquitin E3 ligases.
Cell Res., 24, 2014
|
|
6WJH
 
 | |
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R40
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 87G7 heavy chain variable region, ... | | Authors: | Hurdiss, D.L. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Sci Immunol, 7, 2022
|
|
8WDW
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2, ... | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
5CQ2
 
 | | Crystal Structure of tandem WW domains of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
3L15
 
 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPQ
 
 | |
