7O7Z
 
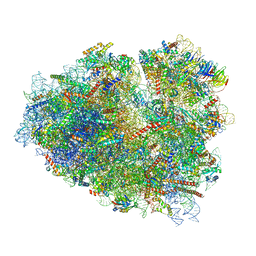 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
6DGV
 
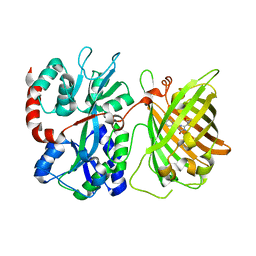 | |
7RWH
 
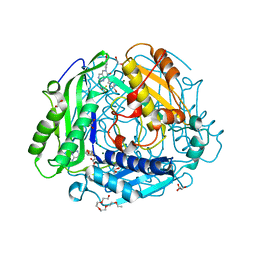 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWG
 
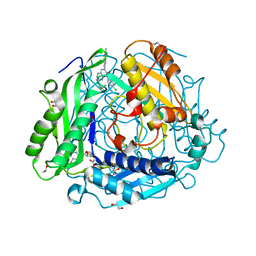 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW7
 
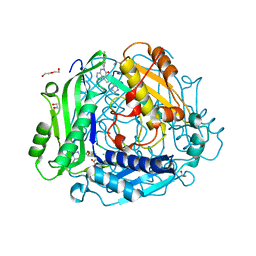 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | Descriptor: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW5
 
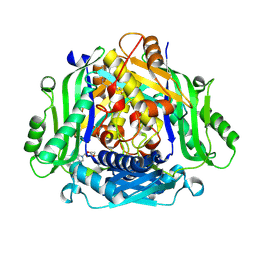 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7NXZ
 
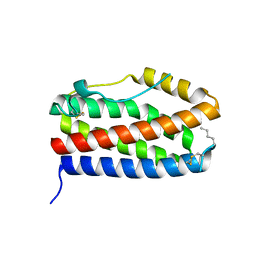 | |
6XVT
 
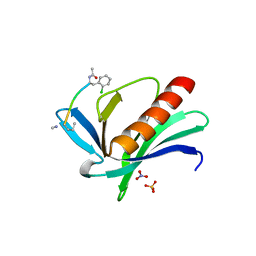 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDDL-NH2 | | Descriptor: | ACY-SC1-SC2-SC3-SC4-SC5-NME, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2NYT
 
 | | The APOBEC2 Crystal Structure and Functional Implications for AID | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Prochnow, C, Bransteitter, R, Klein, M, Goodman, M, Chen, X. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The APOBEC-2 crystal structure and functional implications for the deaminase AID.
Nature, 445, 2007
|
|
6XXR
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEA-NH2 | | Descriptor: | Ac-[2-Cl-F]-PPPPTEDEA-NH2, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FA3
 
 | |
7AQ1
 
 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
1KSR
 
 | | THE REPEATING SEGMENTS OF THE F-ACTIN CROSS-LINKING GELATION FACTOR (ABP-120) HAVE AN IMMUNOGLOBULIN FOLD, NMR, 20 STRUCTURES | | Descriptor: | GELATION FACTOR | | Authors: | Fucini, P, Renner, C, Herberhold, C, Noegel, A.A, Holak, T.A. | | Deposit date: | 1997-02-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold.
Nat.Struct.Biol., 4, 1997
|
|
6ERC
 
 | | Peroxidase A from Dictyostelium discoideum (DdPoxA) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolussi, A, Mlynek, G, Furtmueller, P.G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2017-10-17 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.50001836 Å) | | Cite: | Secreted heme peroxidase from Dictyostelium discoideum: Insights into catalysis, structure, and biological role.
J. Biol. Chem., 293, 2018
|
|
8QMQ
 
 | | Succinic semialdehyde dehydrogenase from E. coli with bound NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMR
 
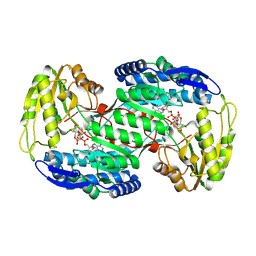 | |
8QMT
 
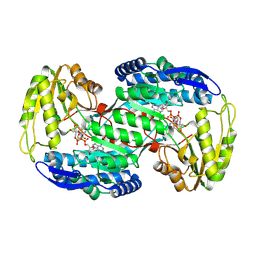 | | Succinic semialdehyde dehydrogenase from E. coli with Q262R substitution and bound NAD+, succinic semialdehyde | | Descriptor: | 4-oxobutanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMS
 
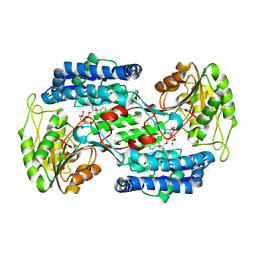 | |
7P4L
 
 | | Crystal structure of the trimeric ectodomain of archaeal Fusexin1 (Fsx1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fusexin1, ... | | Authors: | Nishio, S, Tunyasuvunakool, K, Jumper, J, De Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins.
Nat Commun, 13, 2022
|
|
4EFX
 
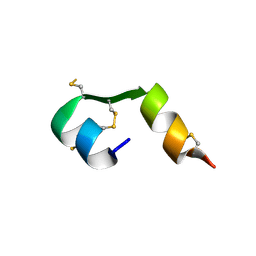 | |
7ZKX
 
 | | SRPK2 IN COMPLEX WITH INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, ... | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
7ZKS
 
 | | SRPK1 IN COMPLEX WITH INHIBITOR | | Descriptor: | CHLORIDE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, SRSF protein kinase 1 | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
5AE0
 
 | |
5EQT
 
 | |
