7OSI
 
 | | ABC Transporter complex NosDFYL, R-domain 3 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSF
 
 | | ABC Transporter complex NosDFYL, R-domain 1 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSH
 
 | | ABC Transporter complex NosDFYL, R-domain 2 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
1H87
 
 | | Gadolinium derivative of tetragonal Hen Egg-White Lysozyme at 1.7 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Girard, E, Chantalat, L, Vicat, J, Kahn, R. | | Deposit date: | 2001-01-25 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Gd-Hp-Do3A, a Complex to Obtain High-Phasing-Power Heavy Atom Derivatives for Sad and MAD Experiments. Results with Tetragonal Hen Egg-White Lysozyme
Acta Crystallogr.,Sect.D, 58, 2001
|
|
6GVI
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidine-4,6-diamine | | Descriptor: | 3-(2-azanyl-1,3-benzoxazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-d]pyrimidine-4,6-diamine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6GVH
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-4-chloro-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-chloranyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
1H80
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
6GVG
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-4-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-methyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5DUP
 
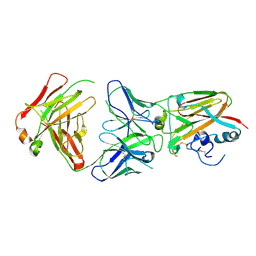 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody AVFluIgG03 | | Descriptor: | AVFluIgG03 Heavy Chain, AVFluIgG03 Light Chain, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUT
 
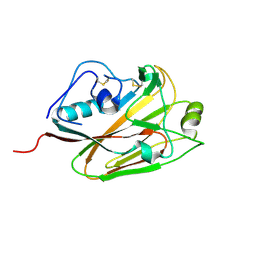 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
2K1Z
 
 | |
1QCR
 
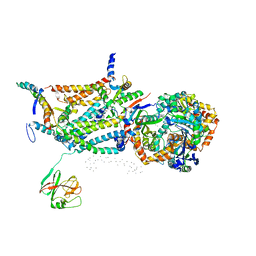 | | CRYSTAL STRUCTURE OF BOVINE MITOCHONDRIAL CYTOCHROME BC1 COMPLEX, ALPHA CARBON ATOMS ONLY | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UBIQUINOL CYTOCHROME C OXIDOREDUCTASE | | Authors: | Xia, D, Yu, C.A, Kim, H, Xia, J.Z, Kachurin, A, Zhang, L, Yu, L, Deisenhofer, J. | | Deposit date: | 1997-05-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria.
Science, 277, 1997
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6F6R
 
 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
7T3K
 
 | | Cryo-EM structure of Csy-AcrIF24 dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7TAW
 
 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7TAX
 
 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3L
 
 | | Cryo-EM structure of Csy-AcrIF24-DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
