7MFB
 
 | | Crystal structure of antibody 10E8v4 Fab - light chain H31F variant | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MFA
 
 | | Crystal structure of antibody 10E8v4-P100fA+P100gA Fab | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF8
 
 | | Crystal structure of antibody 10E8v4-P100fA Fab in space group P6422 | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF7
 
 | | Crystal structure of antibody 10E8v4-P100gA Fab | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF9
 
 | | Crystal structure of antibody 10E8v4-P100fA Fab in space group C2 | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7LSQ
 
 | |
5J2N
 
 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated EFdA-MP at the P-(post-translocation) site and dTMP at the N-(pre-translocation) site | | Descriptor: | DNA (27-MER), DNA 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', MAGNESIUM ION, ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J2Q
 
 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated a mismatched EFdA-MP at the N-(pre-translocation) site | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(dihydrogen phosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(6FM)P*(6FM))-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5WEV
 
 | |
6EE0
 
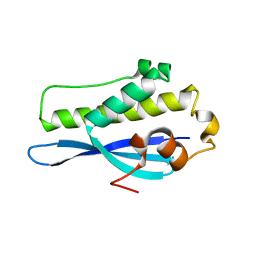 | | Crystal Structure of SNX23 PX domain | | Descriptor: | Kinesin-like protein KIF16B | | Authors: | Chandra, M, Collins, B.M. | | Deposit date: | 2018-08-12 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities.
Nat Commun, 10, 2019
|
|
6ECM
 
 | |
7LUI
 
 | |
7LSM
 
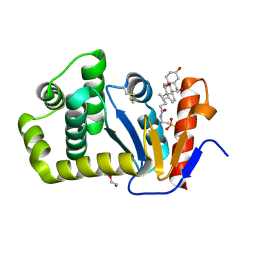 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
7LUH
 
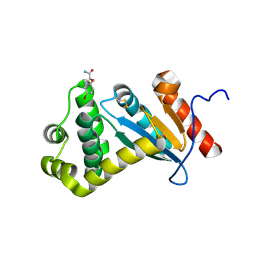 | |
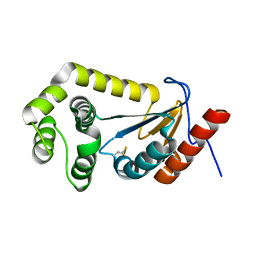
7LUJ
 
 | |
6E51
 
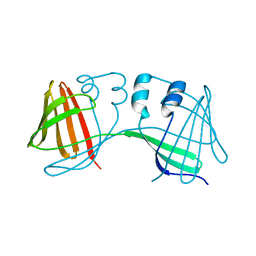 | |
6E50
 
 | |
6E7M
 
 | |
6E5R
 
 | |
5IVL
 
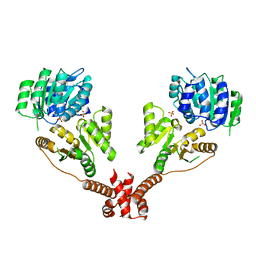 | | CshA Helicase | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA, SULFATE ION | | Authors: | Huen, J, Lin, C.-L, Yi, W.-L, Li, C.-L, Yuan, H. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into a Unique Dimeric DEAD-Box Helicase CshA that Promotes RNA Decay.
Structure, 25, 2017
|
|
6E5S
 
 | |
6E6L
 
 | |
7C01
 
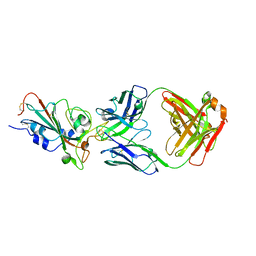 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
4LST
 
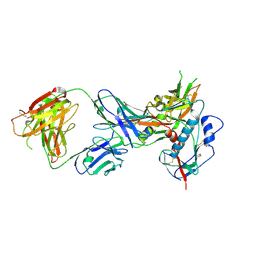 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade C strain ZM176.66 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
6ZNO
 
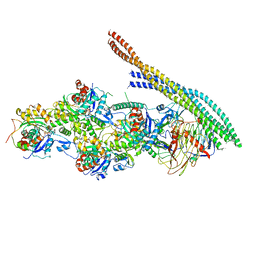 | | The pointed end complex of dynactin with the p150 projection docked | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
