7D1M
 
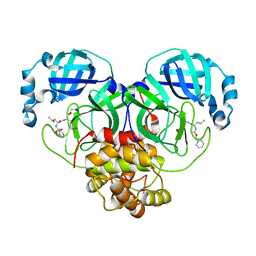 | | CRYSTAL STRUCTURE OF THE SARS-CoV-2 MAIN PROTEASE COMPLEXED WITH GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fu, L.F, Gilski, M, Shabalin, I, Gao, G.F, Qi, J.X. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7C4Y
 
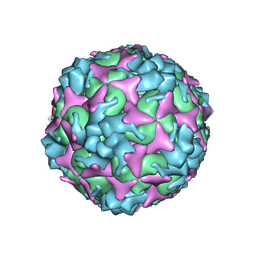 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7EIN
 
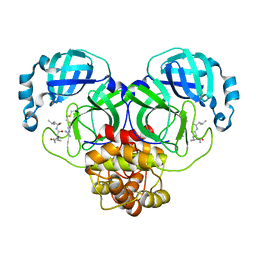 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7EDO
 
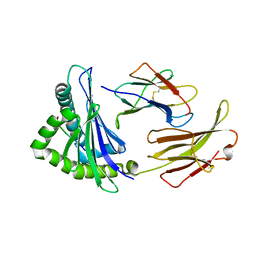 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
7DRV
 
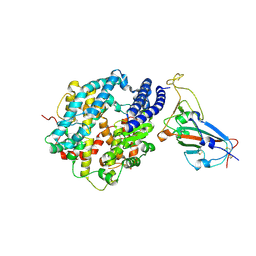 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
7CZE
 
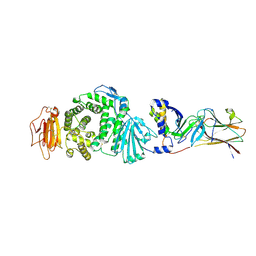 | | Crystal structure of Epstein-Barr virus (EBV) gHgL and in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
7D6I
 
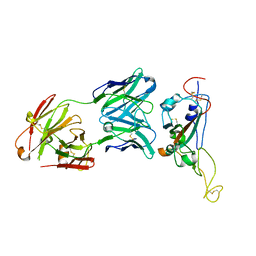 | | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of GH12-Fab, Light chain of GH12-Fab, ... | | Authors: | Shi, R, Qi, J.X, Gao, G.F, Yan, J.H. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2
To Be Published
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CDZ
 
 | |
7C8K
 
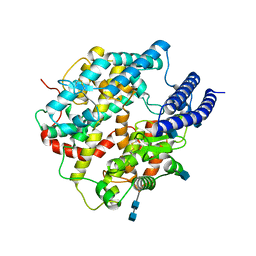 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CE0
 
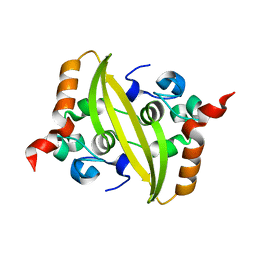 | |
7F9M
 
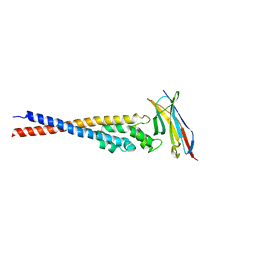 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9K
 
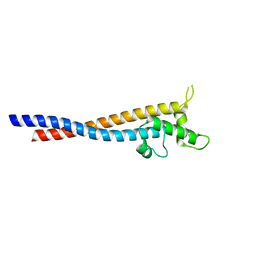 | | Crystal structure of the variable region of Plasmodium RIFIN #6(PF3D7_1400600) | | Descriptor: | Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9N
 
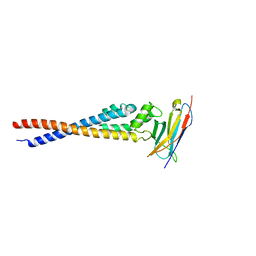 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9L
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #6 (PF3D7_1400600) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6IEA
 
 | | Structure of RVFV Gn and human monoclonal antibody R13 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NSmGnGc, ... | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEB
 
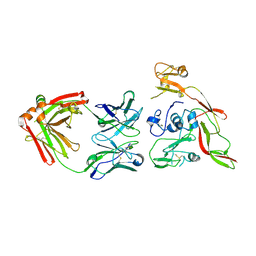 | | Structure of RVFV Gn and human monoclonal antibody R15 | | Descriptor: | NSmGnGc, R15 H chain, R15 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEC
 
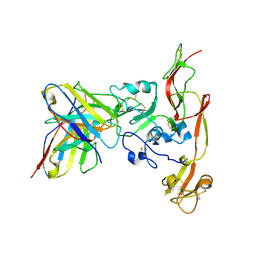 | | Structure of RVFV Gn and human monoclonal antibody R17 | | Descriptor: | NSmGnGc, R17 H chain, R17 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEK
 
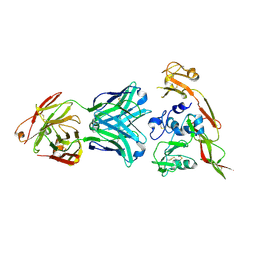 | | Structure of RVFV Gn and human monoclonal antibody R12 | | Descriptor: | Heavy chain of Fab R12, Light chain of Fab R12, NSmGnGc | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6J14
 
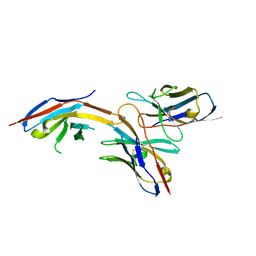 | | Complex structure of GY-14 and PD-1 | | Descriptor: | GY-14 heavy chain V fragment, GY-14 light chain V fragment, Programmed cell death protein 1 | | Authors: | Chen, D, Tan, S, Whang, H, Zhang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
6IEX
 
 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | Authors: | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
6KUR
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
