4LMC
 
 | |
4LM7
 
 | |
7EMN
 
 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7FCP
 
 | |
7FCQ
 
 | |
7WK0
 
 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7X2H
 
 | |
7XD2
 
 | |
7Y7P
 
 | | QDE-1 in complex with RNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7Q
 
 | | QDE-1 in complex with RNA template, RNA primer and 3'-dGTP | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7R
 
 | | QDE-1 in complex with DNA template, RNA primer and 3'-dGTP | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*AP*AP*CP*TP*AP*CP*CP*GP*TP*CP*GP*GP*A)-3'), ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7S
 
 | | QDE-1 in complex with DNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7T
 
 | | QDE-1 in complex with 12nt DNA template, ATP and 3'-dGTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*G*AP*GP*AP*CP*CP*TP*TP*TP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
7E9Q
 
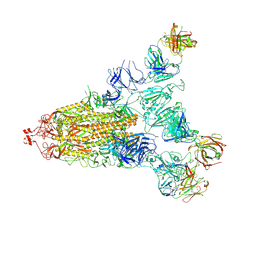 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
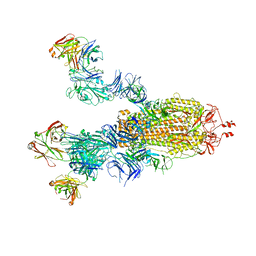 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9P
 
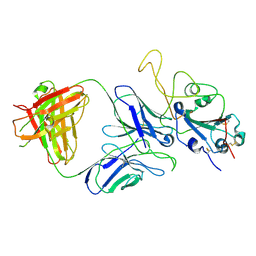 | |
7E9N
 
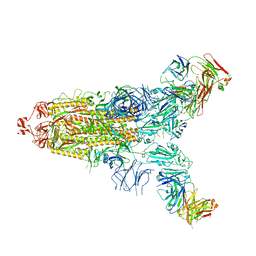 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
