4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MZJ
 
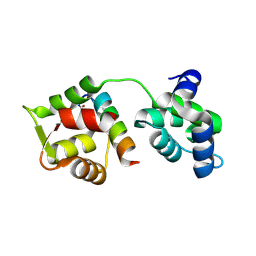 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[801,805], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, Myosin-A | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
1YOK
 
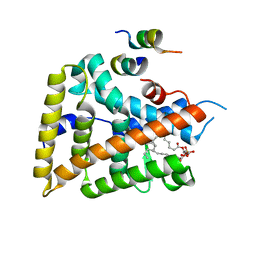 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
1EAL
 
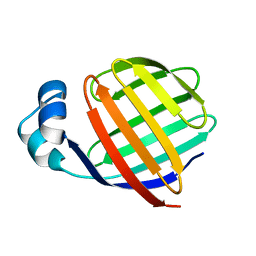 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
1YQV
 
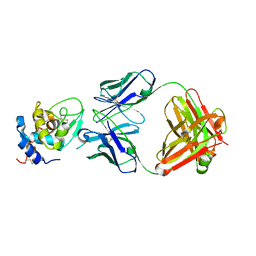 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1YW2
 
 | | Mutated Mus Musculus P38 Kinase (mP38) | | Descriptor: | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Laughlin, S.K, Clark, M.P, Djung, J.F, Golebiowski, A, Brugel, T.A, Sabat, M, Bookland, R.G, Laufersweiler, M.J, Vanrens, J.C, Townes, J.A, De, B, Hsieh, L.C, Xu, S.C, Walter, R.L, Mekel, M.J, Janusz, M.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The development of new isoxazolone based inhibitors of tumor necrosis factor-alpha (TNF-alpha) production.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4CF3
 
 | |
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OGC
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
1ZGO
 
 | |
1ZJP
 
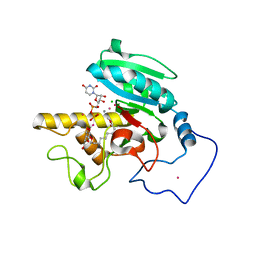 | | Crystal Structure of Human Galactosyltransferase (GTB) Complexed with Galactose-grease | | Descriptor: | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
1I1F
 
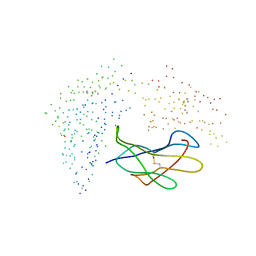 | | Crystal structure of human class i mhc (hla-a2.1) complexed with beta 2-microglobulin and hiv-rt variant peptide i1y | | Descriptor: | PROTEIN (BETA 2-MICROGLOBULIN), PROTEIN (CLASS I HISTOCOMPATIBILITY ANTIGEN, GOGO-A0201 ALPHA CHAIN), ... | | Authors: | Kirksey, T.J, Pogue-Caley, R.R, Frelinger, J.A, Collins, E.J. | | Deposit date: | 1999-04-16 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the increased immunogenicity of two HIV-reverse transcriptase peptide variant/class I major histocompatibility complexes.
J.Biol.Chem., 274, 1999
|
|
1ZGP
 
 | |
1ZI4
 
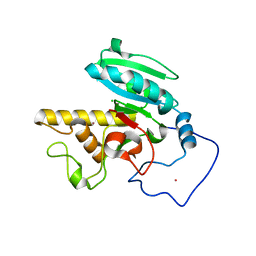 | | Crystal Structure of Human N-acetylgalactosaminyltransferase (GTA) Complexed with H type II Trisaccharide | | Descriptor: | CHLORIDE ION, Histo-blood group ABO system transferase (NAGAT) Includes: Glycoprotein-fucosylgalactoside alpha-N- acetylgalactosaminyltransferase, MERCURY (II) ION, ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
1ZJG
 
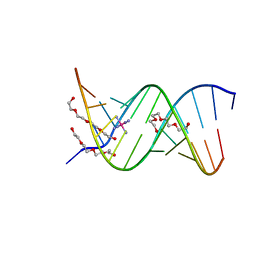 | | 13mer-co | | Descriptor: | 5'-D(*AP*TP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*CP*A)-3', COBALT HEXAMMINE(III), ... | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
4FKI
 
 | |
4FKT
 
 | |
1I72
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-07 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7C
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH METHYLGLYOXAL BIS-(GUANYLHYDRAZONE) | | Descriptor: | 1,4-DIAMINOBUTANE, METHYLGLYOXAL BIS-(GUANYLHYDRAZONE), S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7M
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE | | Descriptor: | 1,4-DIAMINOBUTANE, 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4F11
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor GBR2 | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Geng, Y, Xiong, D, Mosyak, L, Malito, D.L, Kniazeff, J, Chen, Y, Burmakina, S, Quick, M, Bush, M, Javitch, J.A, Pin, J.-P, Fan, Q.R. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2.
Nat.Neurosci., 15, 2012
|
|
1I10
 
 | | HUMAN MUSCLE L-LACTATE DEHYDROGENASE M CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-LACTATE DEHYDROGENASE M CHAIN, ... | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
