6Q2W
 
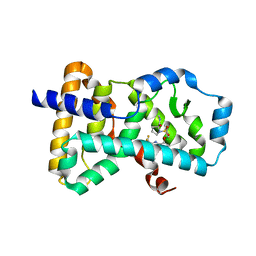 | | Crystal structure of human ROR gamma LBD in complex with a quinoline sulfonamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[4-imidazol-1-yl-2-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]sulfonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Ciesielski, F, Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1ZK4
 
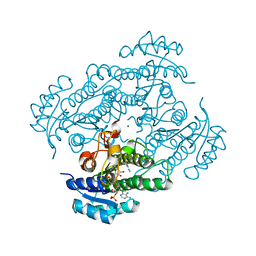 | | Structure of R-specific alcohol dehydrogenase (wildtype) from Lactobacillus brevis in complex with acetophenone and NADP | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ATT
 
 | |
2JRY
 
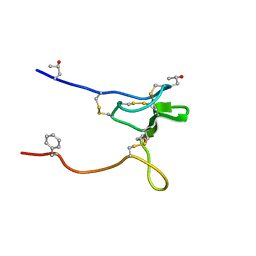 | | Structure and Sodium Channel Activity of an Excitatory I1-Superfamily Conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D, Babon, J, Yang, X, Fiedler, B, Chen, P, Yoshikami, D, Olivera, B, Bulaj, G, Norton, R. | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin.
Biochemistry, 46, 2007
|
|
8PXB
 
 | |
4PVU
 
 | | Crystal structure of the complex between PPARgamma-LBD and the R enantiomer of Mbx-102 (Metaglidasen) | | Descriptor: | (2R)-(4-chlorophenyl)[3-(trifluoromethyl)phenoxy]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Piemontese, L, Lavecchia, A. | | Deposit date: | 2014-03-18 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | On the metabolically active form of metaglidasen: improved synthesis and investigation of its peculiar activity on peroxisome proliferator-activated receptors and skeletal muscles.
Chemmedchem, 10, 2015
|
|
6HBN
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA/CSKN2A1 GENE PRODUCT) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Hochscherf, J, Dimper, V, Witulski, B, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
2JZ2
 
 | | Solution NMR structure of Ssl0352 protein from Synechocystis sp. PCC 6803. Northeast Structural Genomics Consortium target SgR42 | | Descriptor: | Ssl0352 protein | | Authors: | Eletsky, A, Sukumaran, D, Wang, D, Hamilton, K, Foote, E.L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-22 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Ssl0352 protein from Synechocystis sp.
To be Published
|
|
6Q9K
 
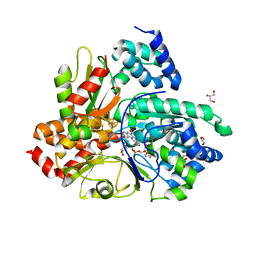 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF S96M bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HPX
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[(3-chlorophenyl)methyl]-1-(2-pyrrolidin-1-ylethyl)benzimidazole-5-carboxamide | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6QB4
 
 | | Mcl1-scFv complex with an indole acid inhibitor | | Descriptor: | 3-[3-[[(1~{R})-1,2,3,4-tetrahydronaphthalen-1-yl]oxy]propyl]-7-(1,3,5-trimethylpyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QB9
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | L(+)-TARTARIC ACID, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1OEV
 
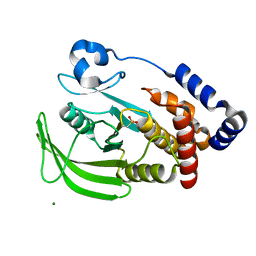 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
6HLA
 
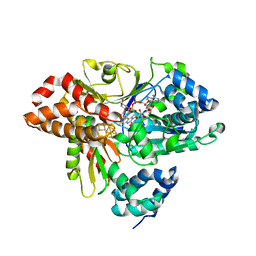 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NADH | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
4QI7
 
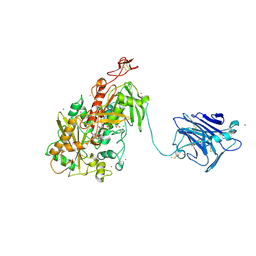 | | Cellobiose dehydrogenase from Neurospora crassa, NcCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
2JWZ
 
 | | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins | | Descriptor: | Ubiquitin | | Authors: | Haririnia, A, Verma, R, Purohit, N, Twarog, M, Deshaies, R, Bolon, D, Fushman, D. | | Deposit date: | 2007-10-31 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins.
J.Mol.Biol., 375, 2008
|
|
2CLM
 
 | | Tryptophan Synthase (external aldimine state) in complex with N-(4'- trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-28 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
2CLO
 
 | | Tryptophan Synthase (external aldimine state) in complex with (naphthalene-2'-sulfonyl)-2-amino-1-ethylphosphate (F19) | | Descriptor: | 2-[(2-NAPHTHYLSULFONYL)AMINO]ETHYL DIHYDROGEN PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-28 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
7ZQ7
 
 | | Structure of RpF-1 | | Descriptor: | Crotonase/enoyl-CoA hydratase family protein | | Authors: | Sanchez-Alba, L, Reverter, D, Conchillo, O, Yero, D, Daura, X, Gibert, I. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of RpF-1
To Be Published
|
|
1YQ3
 
 | | Avian respiratory complex ii with oxaloacetate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Coenzyme Q10, (2Z,6E,10Z,14E,18E,22E,26Z)-isomer, ... | | Authors: | Huang, L, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1NX1
 
 | | Calpain Domain VI Complexed with Calpastatin Inhibitory Domain C (DIC) | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
7M7C
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
8QPH
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra 14 crystals | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | VMXm - A sub-micron focus macromolecular crystallography beamline at Diamond Light Source.
J.Synchrotron Radiat., 31, 2024
|
|
4QI4
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase, MtDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
1ASZ
 
 | | THE ACTIVE SITE OF YEAST ASPARTYL-TRNA SYNTHETASE: STRUCTURAL AND FUNCTIONAL ASPECTS OF THE AMINOACYLATION REACTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Cavarelli, J, Rees, B, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction.
EMBO J., 13, 1994
|
|
