1UUZ
 
 | | IVY:A NEW FAMILY OF PROTEIN | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Lembo, F, Byrne, D, Maza, C, Claverie, J.M. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1UTZ
 
 | | Crystal Structure of MMP-12 complexed to (2R)-3-({[4-[(pyridin-4-yl)phenyl]-thien-2-yl}carboxamido)(phenyl)propanoic acid | | Descriptor: | (2R)-3-({[4-[(PYRIDIN-4-YL)PHENYL]-THIEN-2-YL}CARBOXAMIDO)(PHENYL)PROPANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Novel Non-Peptidic, Non-Zinc Chelating Inhibitors Bound to Mmp-12
J.Mol.Biol., 341, 2004
|
|
1UVN
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 ca2+ inhibition complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UTT
 
 | | Crystal Structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL) ETHYL-4-(4'-ETHOXY [1,1'-BIPHENYL]-4-YL)-4-OXOBUTANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J. Mol. Biol., 341, 2004
|
|
1UOS
 
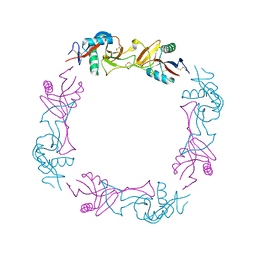 | | The Crystal Structure of the Snake Venom Toxin Convulxin | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Batuwangala, T, Leduc, M, Gibbins, J.M, Bon, C, Jones, E.Y. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Snake-Venom Toxin Convulxin
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3D2Y
 
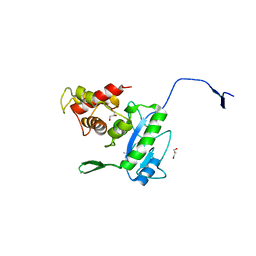 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3E0S
 
 | | Crystal structure of an uncharacterized protein from Chlorobium tepidum | | Descriptor: | SULFATE ION, uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Powell, A, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of an uncharacterized protein from Chlorobium tepidum
To be Published
|
|
3R5U
 
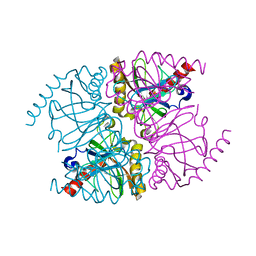 | | The structure of manganese bound Thermococcus thioreducens Inorganic pyrophosphatase | | Descriptor: | MANGANESE (II) ION, Tt-IPPase | | Authors: | Hughes, R.C, Coates, L, Meehan, E.J, Garcia-Ruiz, J.M, Ng, J.D. | | Deposit date: | 2011-03-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of manganese bound Thermococcus thioreducens Inorganic pyrophosphatase
To be Published
|
|
1UVL
 
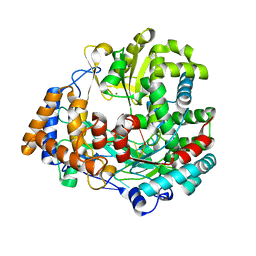 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5nt RNA. Conformation B | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
3RCF
 
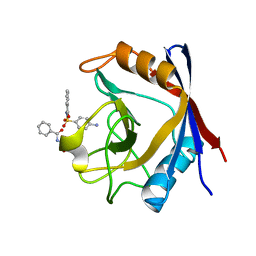 | | Human cyclophilin D complexed with N-[(4-aminophenyl)sulfonyl]benzamide | | Descriptor: | N-[(4-aminophenyl)sulfonyl]benzamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1W1N
 
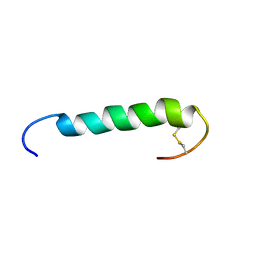 | | The solution structure of the FATC Domain of the Protein Kinase TOR1 from yeast | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE TOR1 | | Authors: | Dames, S.A, Mulet, J.M, Rathgeb-Szabo, K, Hall, M.N, Grzesiek, S. | | Deposit date: | 2004-06-23 | | Release date: | 2005-03-16 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability.
J. Biol. Chem., 280, 2005
|
|
3DLL
 
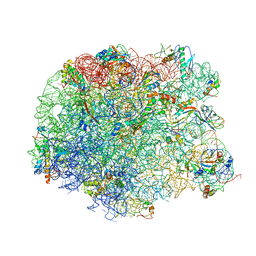 | | The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Starosta, A.L, Connell, S.R, Fucini, P. | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1W2Z
 
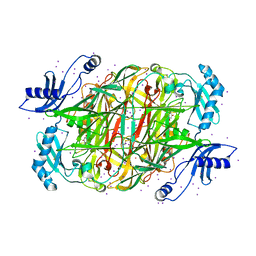 | | PSAO and Xenon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMINE OXIDASE, COPPER CONTAINING, ... | | Authors: | Duff, A.P, Trambaiolo, D.M, Cohen, A.E, Ellis, P.J, Juda, G.A, Shepard, E.M, Langley, D.B, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-Binding Sites in Copper Amine Oxidases.
J.Mol.Biol., 344, 2004
|
|
3DZT
 
 | | AeD7-leukotriene E4 complex | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Andersen, J.F, Calvo, E, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2008-07-30 | | Release date: | 2009-02-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3RCL
 
 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
3DYE
 
 | | Crystal structure of AED7-norepineprhine complex | | Descriptor: | BROMIDE ION, D7 protein, GLYCEROL, ... | | Authors: | Andersen, J.F, Calvo, E, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2008-07-27 | | Release date: | 2009-02-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
3DO9
 
 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
1UUF
 
 | |
1UPC
 
 | | Carboxyethylarginine synthase from Streptomyces clavuligerus | | Descriptor: | CARBOXYETHYLARGININE SYNTHASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Caines, M.E.C, Elkins, J.M, Hewitson, K.S, Schofield, C.J. | | Deposit date: | 2003-09-29 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure and Mechanistic Implications of N2-(2-Carboxyethyl)Arginine Synthase, the First Enzyme in the Clavulanic Acid Biosynthesis Pathway
J.Biol.Chem., 279, 2004
|
|
1UNX
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UW7
 
 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
1UO1
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UQR
 
 | | Type II 3-dehydroquinate dehydratase (DHQase) from Actinobacillus pleuropneumoniae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Maes, D, Gonzalez-Ramirez, L.A, Lopez-Jaramillo, J, Yu, B, De Bondt, H, Zegers, I, Afonina, E, Garcia-Ruiz, J.M, Gulnik, S. | | Deposit date: | 2003-10-16 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Study of the Type II 3-Dehydroquinate Dehydratase from Actinobacillus Pleuropneumoniae
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UO2
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
