6RP3
 
 | | Truncated Norcoclaurine synthase with reaction intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[[(2~{R})-2-phenylpropyl]amino]ethyl]benzene-1,2-diol, S-norcoclaurine synthase | | Authors: | Keep, N.H, Roddan, R, Sula, A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Acceptance and Kinetic Resolution of alpha-Methyl-Substituted Aldehydes by Norcoclaurine Synthases
Acs Catalysis, 2019
|
|
5L90
 
 | |
6TOE
 
 | |
6TO4
 
 | |
8AHB
 
 | |
8AHA
 
 | | rsEGFP2 photoswitched to its off-state at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Mantovanelli, A, Adam, V. | | Deposit date: | 2022-07-21 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Photophysical Studies at Cryogenic Temperature Reveal a Novel Photoswitching Mechanism of rsEGFP2.
J.Am.Chem.Soc., 145, 2023
|
|
3N7X
 
 | |
4C4K
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | 1,2-ETHANEDIOL, OBSCURIN, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
7B5W
 
 | |
2OGZ
 
 | | Crystal structure of DPP-IV complexed with Lilly aryl ketone inhibitor | | Descriptor: | 4-[(3R)-3-{[2-(4-FLUOROPHENYL)-2-OXOETHYL]AMINO}BUTYL]BENZAMIDE, Dipeptidyl peptidase | | Authors: | Timm, D.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of non-covalent dipeptidyl peptidase IV inhibitors which induce a conformational change in the active site.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7B5U
 
 | | RCK_C domain dimer of S.agalactiae BusR in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
7B5Y
 
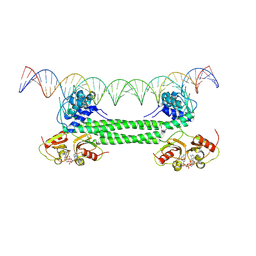 | | S. agalactiae BusR in complex with its busAB-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, BusR binding site in the busAB promotor. strand1, BusR binding site in the busAB promotor. strand2, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
7B5T
 
 | | S. agalactiae BusR transcription factor | | Descriptor: | GntR family transcriptional regulator | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
2LC0
 
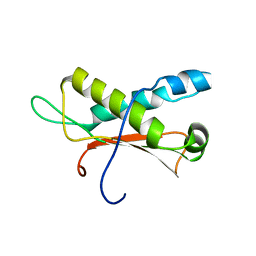 | | Rv0020c_Nter structure | | Descriptor: | Putative uncharacterized protein TB39.8 | | Authors: | Barthe, P.P, Cohen-Gonsaud, M.M, Roumestand, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Mycobacterium tuberculosis Rv0020c Protein and Its Interaction with the PknB Kinase
Structure, 19, 2011
|
|
2LC1
 
 | | Rv0020c_FHA Structure | | Descriptor: | Putative uncharacterized protein TB39.8 | | Authors: | Barthe, P.P, Cohen-Gonsaud, M.M, Roumestand, C.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Mycobacterium tuberculosis Rv0020c Protein and Its Interaction with the PknB Kinase
Structure, 19, 2011
|
|
7Z28
 
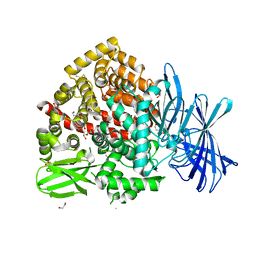 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
5NZZ
 
 | |
5OCM
 
 | | Imine Reductase from Streptosporangium roseum in complex with NADP+ and 2,2,2-trifluoroacetophenone hydrate | | Descriptor: | 2,2,2-trifluoromethyl acetophenone hydrate, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sharma, M, Nestl, B, Grogan, G. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | New imine-reducing enzymes from beta-hydroxyacid dehydrogenases by single amino acid substitutions.
Protein Eng. Des. Sel., 31, 2018
|
|
