7BJ8
 
 | | Structure of L1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
2MJH
 
 | |
5LCQ
 
 | |
5LCR
 
 | |
5MWF
 
 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MVX
 
 | | Human DLL4 C2-EGF3 | | Descriptor: | Delta-like protein 4, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MW7
 
 | | Human Jagged2 C2-EGF3 | | Descriptor: | Protein jagged-2, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MWB
 
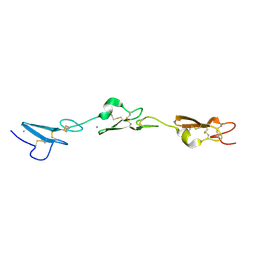 | | Human Notch-2 EGF11-13 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 2, alpha-L-fucopyranose, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MW5
 
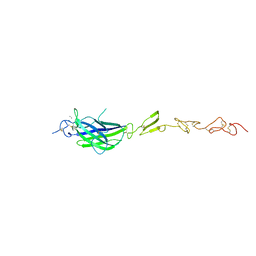 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein jagged-2 | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5GXG
 
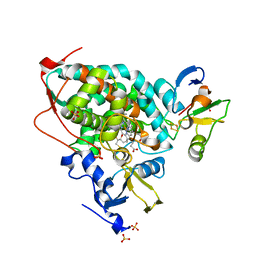 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3LKA
 
 | |
5LCP
 
 | |
5LCT
 
 | |
5LCU
 
 | |
4DO3
 
 | | Structure of FAAH with a non-steroidal anti-inflammatory drug | | Descriptor: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CHLORIDE ION, CYCLOHEXANE AMINOCARBOXYLIC ACID, ... | | Authors: | Garau, G. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Binding Site for Nonsteroidal Anti-inflammatory Drugs in Fatty Acid Amide Hydrolase.
J.Am.Chem.Soc., 135, 2013
|
|
3NXA
 
 | | X-ray structure of the apo form of human S100A16 | | Descriptor: | Protein S100-A16 | | Authors: | Calderone, V. | | Deposit date: | 2010-07-13 | | Release date: | 2010-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human S100A16, a low-affinity calcium binder.
J.Biol.Inorg.Chem., 16, 2011
|
|
6I57
 
 | |
6HIQ
 
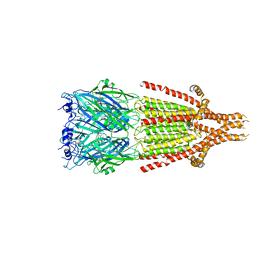 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6IAV
 
 | |
6HIN
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, F conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIO
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIS
 
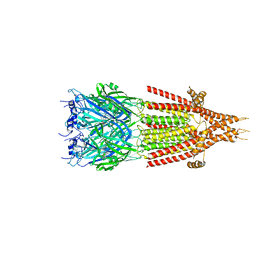 | | Mouse serotonin 5-HT3 receptor, tropisetron-bound, T conformation | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
1DCJ
 
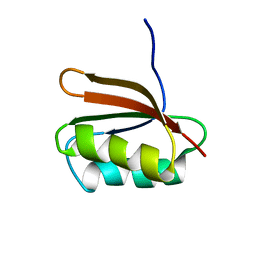 | | SOLUTION STRUCTURE OF YHHP, A NOVEL ESCHERICHIA COLI PROTEIN IMPLICATED IN THE CELL DIVISION | | Descriptor: | YHHP PROTEIN | | Authors: | Katoh, E, Hatta, T, Shindo, H, Mizuno, T, Yamazaki, T. | | Deposit date: | 1999-11-05 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division.
J.Mol.Biol., 304, 2000
|
|
1W6K
 
 | | Structure of human OSC in complex with Lanosterol | | Descriptor: | LANOSTEROL, LANOSTEROL SYNTHASE, octyl beta-D-glucopyranoside | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
1W6J
 
 | | Structure of human OSC in complex with Ro 48-8071 | | Descriptor: | LANOSTEROL SYNTHASE, TETRADECANE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE, ... | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
