1BGP
 
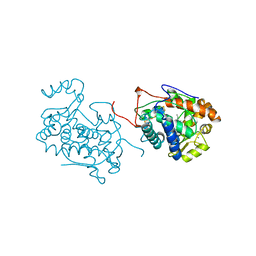 | | CRYSTAL STRUCTURE OF BARLEY GRAIN PEROXIDASE 1 | | Descriptor: | BARLEY GRAIN PEROXIDASE, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Henriksen, A. | | Deposit date: | 1997-07-24 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of barley grain peroxidase refined at 1.9-A resolution. A plant peroxidase reversibly inactivated at neutral pH.
J.Biol.Chem., 273, 1998
|
|
4U67
 
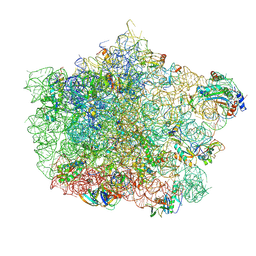 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 | | Descriptor: | 23s RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
7B6A
 
 | | BK Polyomavirus VP1 pentamer core (residues 30-299) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Osipov, E.M, Munawar, A, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
5JNC
 
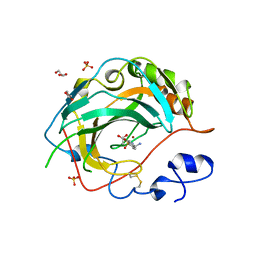 | | Crystal structure for the complex of human carbonic anhydrase IV and 4-aminomethylbenzene sulfonamide | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
1BII
 
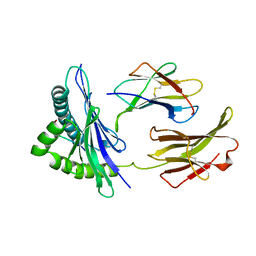 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | Descriptor: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | Authors: | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | Deposit date: | 1998-06-11 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
7C24
 
 | | Glycosidase F290Y at pH8.0 | | Descriptor: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C25
 
 | | Glycosidase Wild Type at pH8.0 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
 | | Glycosidase Wild Type at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
8DVC
 
 | | Receptor ShHTL5 from Striga hermonthica in complex with strigolactone agonist GR24 | | Descriptor: | (3R,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methyl)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Arellano-Saab, A, Skarina, T, Yim, V, Savchenko, A, Stogios, P.J, McCourt, P. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Structural analysis of a hormone-bound Striga strigolactone receptor.
Nat.Plants, 9, 2023
|
|
7C27
 
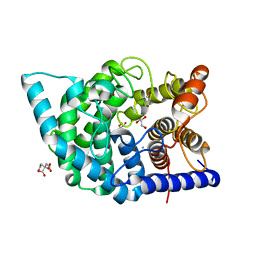 | | Glycosidase F290Y at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
4UCX
 
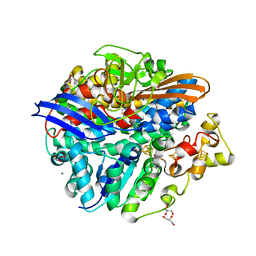 | | Structure of the T18G small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase.
J. Biol. Chem., 290, 2015
|
|
5JN9
 
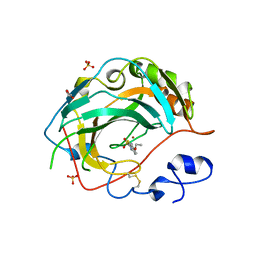 | | Crystal structure for the complex of human carbonic anhydrase IV and ethoxyzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
8DWR
 
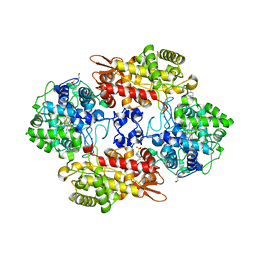 | | Crystal structure of the L333V variant of catalase-peroxidase from Mycobacterium tuberculosis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Diaz-Vilchis, A, Uribe-Vazquez, B, Avila-Linares, A, Rudino-Pinera, E, Soberon, X. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a catalase-peroxidase variant (L333V-KatG) identified in an INH-resistant Mycobacterium tuberculosis clinical isolate.
Biochem Biophys Rep, 37, 2024
|
|
4UCW
 
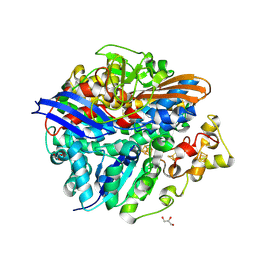 | | Structure of the T18V small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Threonine Stabilizes the Nic and Nir Catalytic Intermediates of [Nife]-Hydrogenase.
J.Biol.Chem., 290, 2015
|
|
1BS8
 
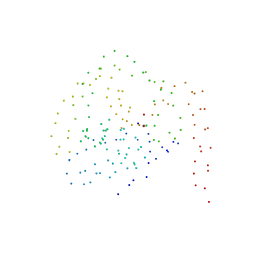 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
7YCA
 
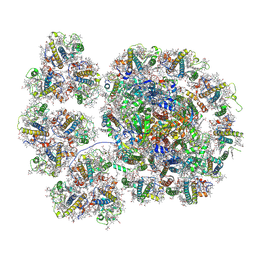 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
8FCI
 
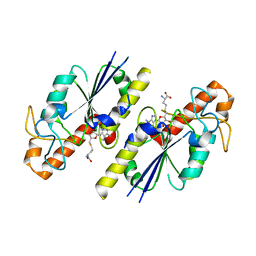 | | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lavallee, T, Mehta, K, Young, A, Cortes, J, Isaacson, B, Brylewski, J, Schlegel, F, Doti, L, Battaile, K, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine
To Be Published
|
|
8FCH
 
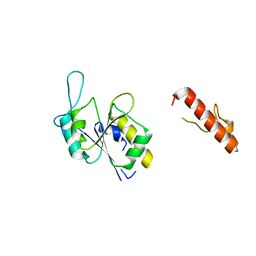 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
4UPG
 
 | |
4UIQ
 
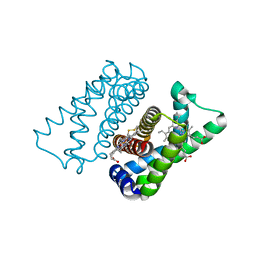 | | Isolated globin domain of the Bordetella pertussis globin-coupled sensor with a heme at the dimer interface | | Descriptor: | GLOBIN-COUPLED SENSOR WITH DIGUANYLATE CYCLASE ACTIVITY, GLYCEROL, IMIDAZOLE, ... | | Authors: | Germani, F, Donne, J, De Schutter, A, Pesce, A, Berghmans, H, Van Hauwaert, M.-L, Moens, L, Van Doorslaer, S, Bolognesi, M, Nardini, M, Dewilde, S. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of a Putative Allosteric Site in the Sensor Domain of Bordetella Pertussis Globin- Coupled Sensor for the Control of Biofilm Formation
To be Published
|
|
4UOU
 
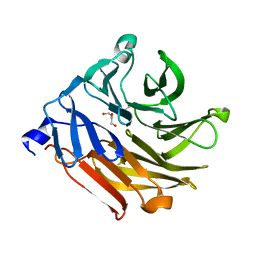 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) - apo-form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA | | Authors: | Houser, J, Komarek, J, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7BF1
 
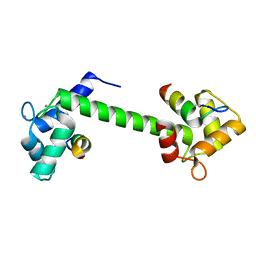 | | Ca2+-Calmodulin in complex with peptide from brain-type creatine kinase in extended 1:2 binding mode | | Descriptor: | ACETYL GROUP, CALCIUM ION, Calmodulin-1, ... | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
7BF2
 
 | | Ca2+-Calmodulin in complex with human muscle form creatine kinase peptide in extended 1:2 binding mode | | Descriptor: | CALCIUM ION, Calmodulin-1, Creatine kinase M-type | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
