7VFU
 
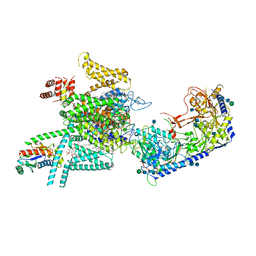 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to ziconotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFV
 
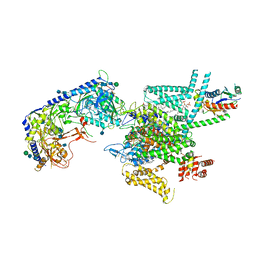 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to PD173212 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(~{tert}-butylamino)-1-oxidanylidene-3-(4-phenylmethoxyphenyl)propan-2-yl]-2-[(4-~{tert}-butylphenyl)methyl-methyl-amino]-4-methyl-pentanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFW
 
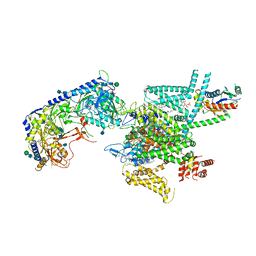 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to CaV2.2-blocker1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(4-chlorophenyl)-1-(2-methoxyphenyl)-3-(2,2,6,6-tetramethyloxan-4-yl)pyrazole, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFS
 
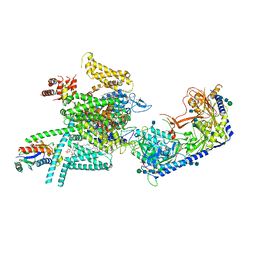 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VKB
 
 | | Crystal structure of the a bacterial kinase complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, HipA (Persistence to inhibition of murein or DNA biosynthesis), HipA_C domain-containing protein, ... | | Authors: | Zhen, X, Ouyang, S.Y. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila.
Nat Commun, 13, 2022
|
|
7VKC
 
 | | Crystal structure of a bacterial Ser/Thr kinase | | Descriptor: | HipA_C domain-containing protein | | Authors: | Zhen, X, Ouyang, S.Y. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila.
Nat Commun, 13, 2022
|
|
7WD8
 
 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCF
 
 | | Crystal structure of the kinase with AMP-PNP | | Descriptor: | HipA_C domain-containing protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Ouyang, S.Y, Zhen, X. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3636 Å) | | Cite: | Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila.
Nat Commun, 13, 2022
|
|
7W54
 
 | | Crystal structure of a bacterial OTU DUB with Ub-PA | | Descriptor: | Lpg2248, Polyubiquitin-B, prop-2-en-1-amine | | Authors: | Zhen, X, Ouyang, S.Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-08-24 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the dual catalytic activity of the Legionella pneumophila ovarian tumor (OTU) domain deubiquitinase LotA.
J.Biol.Chem., 298, 2022
|
|
8JU6
 
 | | Structure of human TRPV4 with antagonist GSK279 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JU5
 
 | | Structure of human TRPV4 with antagonist A1 | | Descriptor: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
7VX1
 
 | | SARS-CoV-2 Beta variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX9
 
 | | SARS-CoV-2 Kappa variant spike protein in complex wth ACE2, state C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXA
 
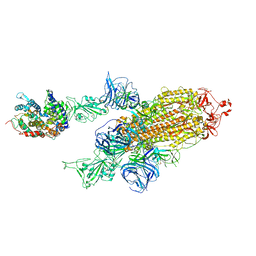 | | SARS-CoV-2 Kappa variant spike protein in complex with ACE2, state C2a | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX5
 
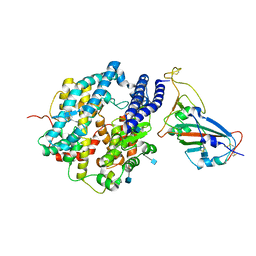 | | ACE2-RBD in SARS-CoV-2 Kappa variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXD
 
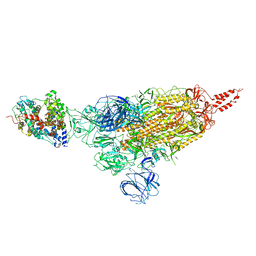 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C1 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX4
 
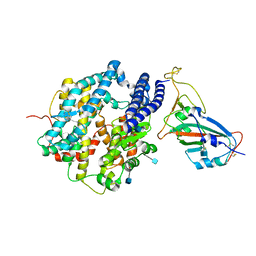 | | ACE2-RBD in SARS-CoV-2 Beta variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXF
 
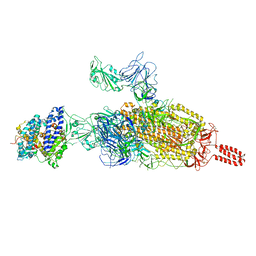 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2B state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXE
 
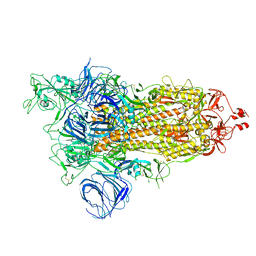 | | SARS-CoV-2 Kappa variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
8K9B
 
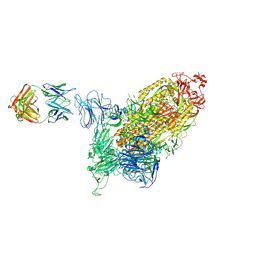 | | SARS-CoV-2 spike protein in complex with one S2H5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of S2H5 Fab, Light chain of S2H5 Fab, ... | | Authors: | Hong, Q, Cong, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis of SARS-CoV-2 immune evasion of NTD neutralizing antibodies.
Dian Zi Xian Wei Xue Bao, 2024
|
|
8K9M
 
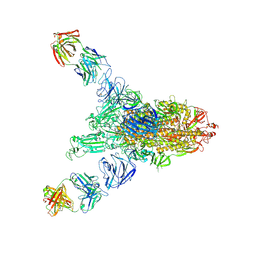 | |
8K9J
 
 | |
7WK2
 
 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
