5MUB
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (CG05) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, triple-helical peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,triple-helical peptide containing the citrullinated C1 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5FGP
 
 | | Crystal structure of D. melanogaster Pur-alpha repeat I-II in complex with DNA. | | Descriptor: | CG1507-PB, isoform B, CHLORIDE ION, ... | | Authors: | Weber, J, Janowski, R, Niessing, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.
Elife, 5, 2016
|
|
7BAI
 
 | | Structure of RIG-I CTD (I875A) bound to p-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*(GDP)P*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
3MES
 
 | | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, DECAMETHONIUM ION, ... | | Authors: | Qiu, W, Wernimont, A, Hills, T, Lew, J, Artz, J.D, Xiao, T, Allali-Hassani, A, Vedadi, M, Kozieradzki, I, Cossar, D, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Hui, R, Ma, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030
TO BE PUBLISHED
|
|
7BAH
 
 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BRZ
 
 | | SOLUTION STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, NMR, 43 STRUCTURES | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-03-12 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
8AQU
 
 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
8AQT
 
 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
1BSZ
 
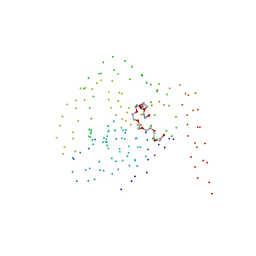 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
5AOU
 
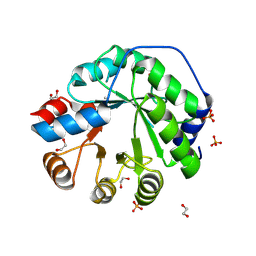 | | Structure of the engineered retro-aldolase RA95.5-8F apo | | Descriptor: | 1,2-ETHANEDIOL, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Obexer, R, Mittl, P, Hilvert, D. | | Deposit date: | 2015-09-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
8AQW
 
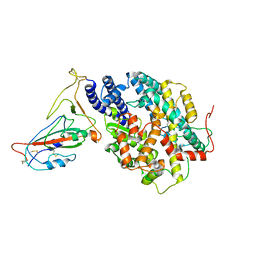 | | BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
6HMQ
 
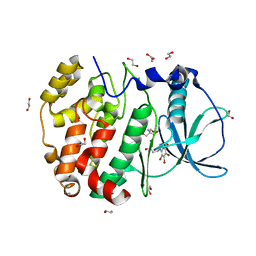 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
3NFA
 
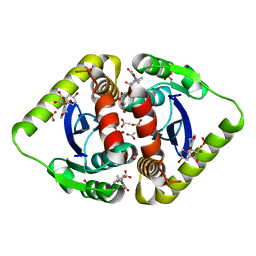 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 6-[(5-bromo-2,3-dioxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF8
 
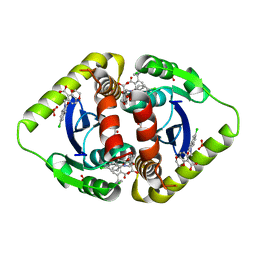 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 6-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
6HV2
 
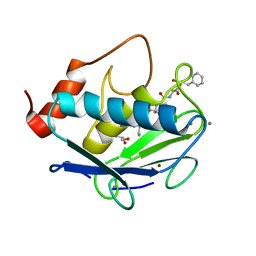 | | MMP-13 in complex with the peptide IMISF | | Descriptor: | CALCIUM ION, Collagenase 3, GLYCEROL, ... | | Authors: | Mittl, P, Riedl, R, Hohl, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6HMB
 
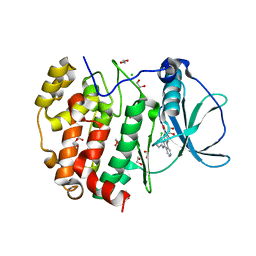 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
1BY5
 
 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
1FMD
 
 | | THE STRUCTURE AND ANTIGENICITY OF A TYPE C FOOT-AND-MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Lea, S, Fry, E, Stuart, D. | | Deposit date: | 1994-02-10 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure and antigenicity of a type C foot-and-mouth disease virus.
Structure, 2, 1994
|
|
1IFX
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH TWO MOLECULES DEAMIDO-NAD | | Descriptor: | NH(3)-DEPENDENT NAD(+) SYNTHETASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2001-04-13 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
