6WDN
 
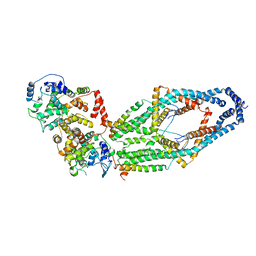 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in low Ca2+ | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | Deposit date: | 2019-03-26 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
3DJD
 
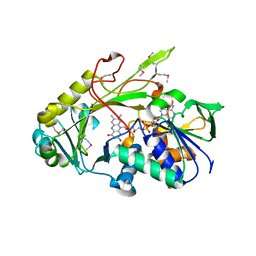 | | Crystal structure of the deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (Amadoriase II) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | Authors: | Collard, F, Zhang, J, Nemet, I, Qanungo, K.R, Monnier, V.M, Yee, V.C. | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the deglycating enzyme fructosamine oxidase (FAOX-II)
To be Published
|
|
6WDO
 
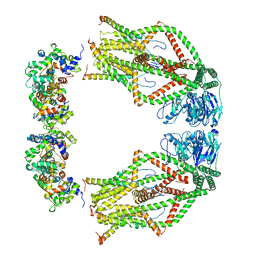 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
3DJE
 
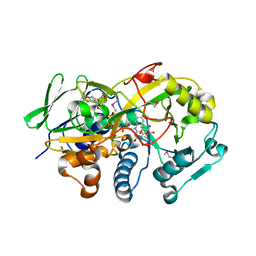 | | Crystal structure of the deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (Amadoriase II) in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Collard, F, Zhang, J, Nemet, I, Qanungo, K.R, Monnier, V.M, Yee, V.C. | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the deglycating enzyme fructosamine oxidase (FAOX-II)
To be Published
|
|
3B9J
 
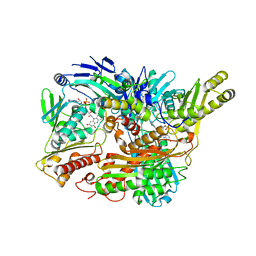 | | Structure of Xanthine Oxidase with 2-hydroxy-6-methylpurine | | Descriptor: | 6-methyl-3,9-dihydro-2H-purin-2-one, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Pauff, J.M, Zhang, J, Bell, C.E, Hille, R. | | Deposit date: | 2007-11-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine.
J.Biol.Chem., 283, 2008
|
|
3C1P
 
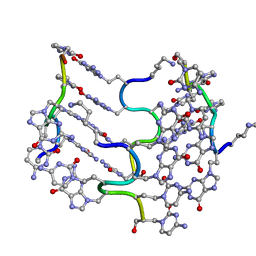 | | Crystal Structure of an alternating D-Alanyl, L-Homoalanyl PNA | | Descriptor: | Peptide Nucleic Acid DLY-HGL-AGD-LHC-AGD-LHC-CUD-LYS | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M, Zhang, J, Diederichsen, U. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Continuous beta-turn fold of an alternating alanyl/homoalanyl peptide nucleic acid.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2L6K
 
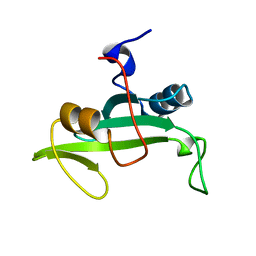 | | Solution Structure of a Nonphosphorylated Peptide Recognizing Domain | | Descriptor: | Tensin-like C1 domain-containing phosphatase | | Authors: | Dai, K, Liao, S, Zhang, J, Zhang, X, Tu, X. | | Deposit date: | 2010-11-22 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1
Plos One, 6, 2011
|
|
8GJM
 
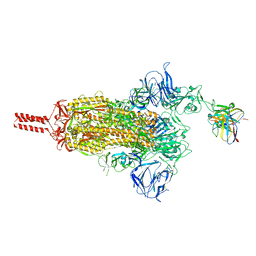 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
3ZDB
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, di-magnesium and potassium | | Descriptor: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDE
 
 | | Potassium free structure of E. coli ExoIX | | Descriptor: | PROTEIN XNI | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDC
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, potassium and calcium | | Descriptor: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDD
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov6 oligonucleotide and potassium | | Descriptor: | 1,2-ETHANEDIOL, 5OV6 DNA, ISOPROPYL ALCOHOL, ... | | Authors: | Flemming, C.S, Hemsworth, G.R, Anstey-Gilbert, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Escherichia coli ExoIX--implications for DNA binding and catalysis in flap endonucleases.
Nucleic Acids Res., 41, 2013
|
|
3ZD8
 
 | | Potassium bound structure of E. coli ExoIX in P1 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZD9
 
 | | Potassium bound structure of E. coli ExoIX in P21 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
3ZDA
 
 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | Descriptor: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
