3WRM
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
3WRJ
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3WN7
 
 | | Crystal Structure of Keap1 in Complex with the N-terminal region of the Nrf2 transcription factor | | Descriptor: | ACETATE ION, Kelch-like ECH-associated protein 1, Peptide from Nuclear factor erythroid 2-related factor 2 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Ohuchi, N, Yamamoto, M. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1
Mol.Cell.Biol., 34, 2014
|
|
3WRL
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
3WRK
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of P450cam intermedite
To be Published
|
|
1RLS
 
 | | CRYSTAL STRUCTURE OF RNASE T1 COMPLEXED WITH THE PRODUCT NUCLEOTIDE 3'-GMP. STRUCTURAL EVIDENCE FOR DIRECT INTERACTION OF HISTIDINE 40 AND GLUTAMIC ACID 58 WITH THE 2'-HYDROXYL GROUP OF RIBOSE | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Gohda, K, Oka, K.-I, Tomita, K.-I, Hakoshima, T. | | Deposit date: | 1994-03-29 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of RNase T1 complexed with the product nucleotide 3'-GMP. Structural evidence for direct interaction of histidine 40 and glutamic acid 58 with the 2'-hydroxyl group of the ribose.
J.Biol.Chem., 269, 1994
|
|
3WA0
 
 | | Crystal structure of merlin complexed with DCAF1/VprBP | | Descriptor: | Merlin, Protein VPRBP | | Authors: | Mori, T, Gotoh, S, Shirakawa, M, Hakoshima, T. | | Deposit date: | 2013-04-20 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DDB1-and-Cullin 4-associated Factor 1 (DCAF1) recognition by merlin/NF2 and its implication in tumorigenesis by CD44-mediated inhibition of merlin suppression of DCAF1 function.
Genes Cells, 19, 2014
|
|
1UL1
 
 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
3X3H
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
1UM7
 
 | |
3WRH
 
 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of P450cam intermedite
To be Published
|
|
3WVL
 
 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
3AIB
 
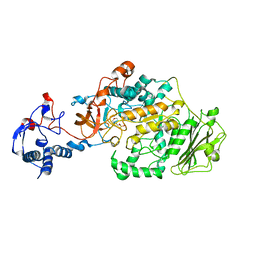 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3X23
 
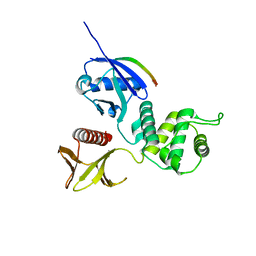 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
3A9H
 
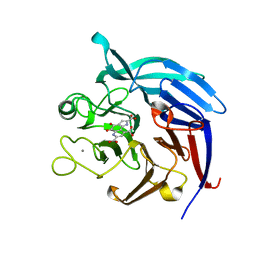 | | Crystal Structure of PQQ-dependent sugar dehydrogenase holo-form | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, Putative uncharacterized protein, ... | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
3AIE
 
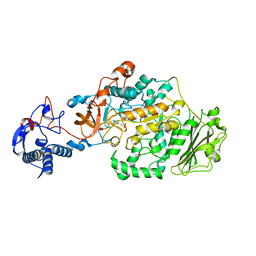 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3A9G
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase apo-form | | Descriptor: | CALCIUM ION, Putative uncharacterized protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
3AIC
 
 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3A9W
 
 | | Crystal structure of L-Threonine bound L-Threonine dehydrogenase (Y137F) from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
3A4V
 
 | | Crystal structure of pyruvate bound L-Threonine dehydrogenase from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
3VTF
 
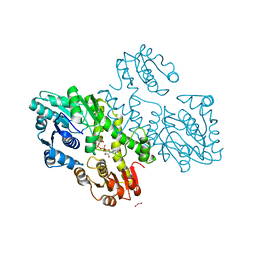 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3W31
 
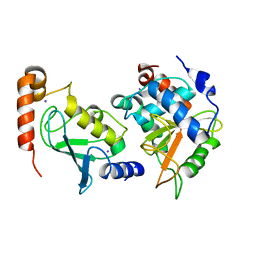 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W30
 
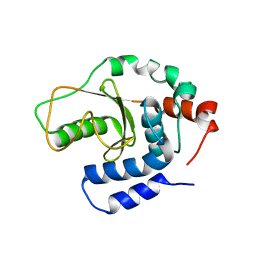 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | ORF169b | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W6Z
 
 | |
