2VNJ
 
 | |
2VNI
 
 | |
2WC1
 
 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
2VNK
 
 | |
2VNH
 
 | |
2E3J
 
 | |
2O98
 
 | | Structure of the 14-3-3 / H+-ATPase plant complex | | Descriptor: | 14-3-3-like protein C, FUSICOCCIN, Plasma membrane H+ ATPase, ... | | Authors: | Ottmann, C, Weyand, M, Wittinghofer, A, Oecking, C. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy
Mol.Cell, 25, 2007
|
|
4F4P
 
 | | SYK in COMPLEX WITH LIGAND LASW836 | | Descriptor: | N-{6-[3-(piperazin-1-yl)phenyl]pyridin-2-yl}-4-(trifluoromethyl)pyridin-2-amine, SULFATE ION, Tyrosine-protein kinase SYK | | Authors: | Lopez, M, Segarra, V, Vidal, B, Wenzkowski, C, Jestel, A, Krapp, S, Blaesse, M, Nagel, S, Schreiner, P. | | Deposit date: | 2012-05-11 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Highly potent aminopyridines as Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5J20
 
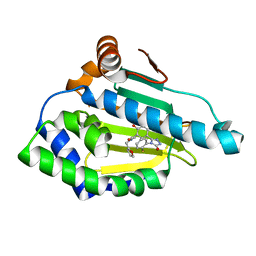 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
2V5U
 
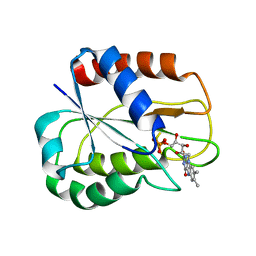 | | I92A FLAVODOXIN FROM ANABAENA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Martinez-Julvez, M, Herguedas, B, Frago, S, Serrano, A, Molina, R, Hamiaux, C, Schierbeek, B, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
5J2X
 
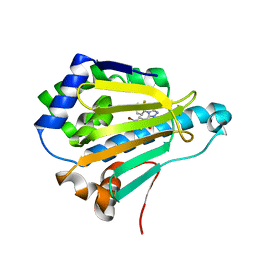 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J8U
 
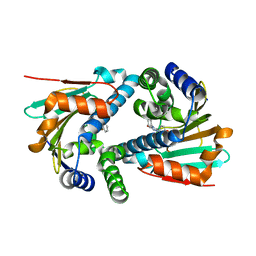 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J82
 
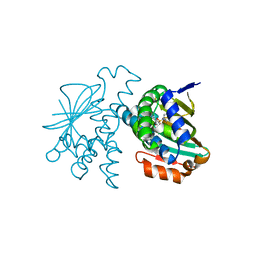 | | Crystal Structure of Hsp90-alpha N-domain in complex 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-isopropyl-N-methyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-(propan-2-yl)benzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J80
 
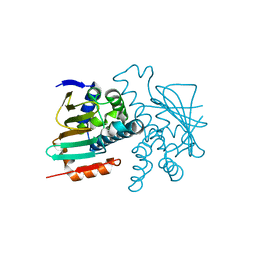 | |
5J6L
 
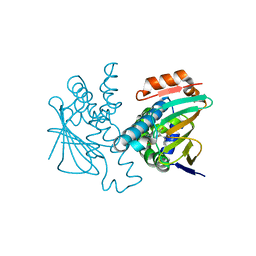 | | Crystal Structure of Hsp90-alpha N-domain in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J2V
 
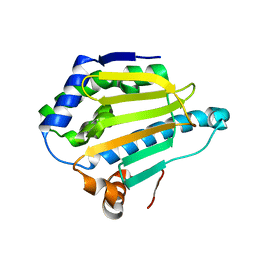 | | Crystal Structure of Hsp90-alpha Apo N-domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J64
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(2,4-dihydroxyphenyl)-4-(2-fluorophenyl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J86
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with 2,4-Dihydroxy-N-methyl-5-(5-oxo-4-o-tolyl-4,5-dihydro-1H-[1,2,4]triazol-3-yl)-N-thiophen-2-ylmethyl-benzamide | | Descriptor: | 2,4-dihydroxy-N-methyl-5-[4-(2-methylphenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(thiophen-2-yl)methyl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J6M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107 mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J8M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J27
 
 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J9X
 
 | | HSP90 in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-11 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J6N
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
6HZV
 
 | | HUMAN JAK3 IN COMPLEX WITH LASW959 PROTEIN IN COMPLEX WITH LIGAND | | Descriptor: | 3-[7-(2-hydroxyethyl)-9-(oxan-4-yl)-8-oxidanylidene-purin-2-yl]imidazo[1,2-a]pyridine-6-carbonitrile, Tyrosine-protein kinase JAK3 | | Authors: | Lozoya, E, Segarra, V, Bach, J, Jestel, A, Lammens, A, Blaesse, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Identification of 2-Imidazopyridine and 2-Aminopyridone Purinones as Potent Pan-Janus Kinase (JAK) Inhibitors for the Inhaled Treatment of Respiratory Diseases.
J.Med.Chem., 62, 2019
|
|
6HZU
 
 | | HUMAN JAK1 IN COMPLEX WITH LASW1393 | | Descriptor: | 2-[4-[8-oxidanylidene-2-[(~{E})-(2-oxidanylidenepyridin-3-ylidene)amino]-7~{H}-purin-9-yl]cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Lozoya, E, Segarra, V, Bach, J, Jestel, A, Lammens, A, Blaesse, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of 2-Imidazopyridine and 2-Aminopyridone Purinones as Potent Pan-Janus Kinase (JAK) Inhibitors for the Inhaled Treatment of Respiratory Diseases.
J.Med.Chem., 62, 2019
|
|
