9BPG
 
 | |
2OOK
 
 | |
2Q3L
 
 | |
3K5J
 
 | |
8ICU
 
 | |
8ICX
 
 | |
8ICV
 
 | |
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
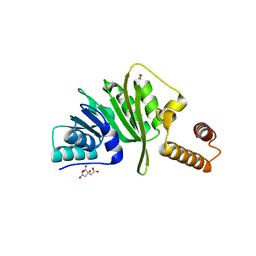 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
3CM1
 
 | |
3DEE
 
 | |
4D4Q
 
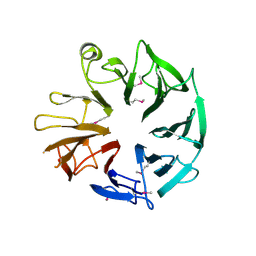 | | Crystal Structure of Kti13/AtS1 | | Descriptor: | PROTEIN ATS1 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
2ICH
 
 | |
5X7V
 
 | |
5X5G
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
2IAY
 
 | |
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.106 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
8RMJ
 
 | | Drosophila Semaphorin 2b in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nourisanami, F, Sobol, M, Rozbesky, D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanisms of proteoglycan-mediated semaphorin signaling in axon guidance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5EVO
 
 | | Structure of Dehydroascrobate Reductase from Pennisetum Americanum in complex with two non-native ligands, Acetate in the G-site and Glycerol in the H-site | | Descriptor: | ACETATE ION, Dehydroascorbate reductase, GLYCEROL | | Authors: | Kumar, A.O, Das, B.K, Arockiasamy, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
7DK1
 
 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
