1M33
 
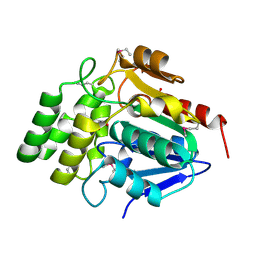 | | Crystal Structure of BioH at 1.7 A | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-PROPANOIC ACID, BioH protein | | Authors: | Sanishvili, R, Savchenko, A, Skarina, T, Edwards, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-26 | | Release date: | 2003-01-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
1Z77
 
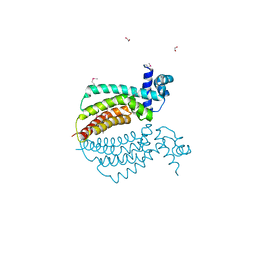 | | Crystal structure of transcriptional regulator protein from Thermotoga maritima. | | Descriptor: | 1,2-ETHANEDIOL, transcriptional regulator (TetR family) | | Authors: | Koclega, K.D, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcriptional regulator TM1030 from Thermotoga maritima solved by an unusual MAD experiment.
J.Struct.Biol., 159, 2007
|
|
1YOY
 
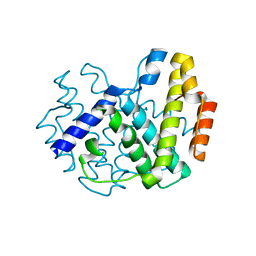 | |
5KOL
 
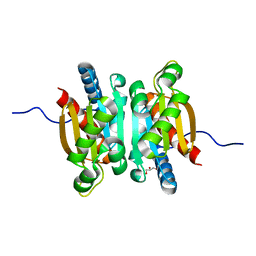 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECK1530/EC0983 from Escherichia coli | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-30 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | To be published
To Be Published
|
|
5KVK
 
 | | Crystal structure of the Competence-Damaged Protein (CinA) Superfamily Protein KP700603 from Klebsiella pneumoniae 700603 | | Descriptor: | Protein KP700603 | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | To be published
To Be Published
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
7MX5
 
 | | Crystal structure of TolB from Acinetobacter baumannii | | Descriptor: | Tol-Pal system protein TolB | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of TolB from Acinetobacter baumannii
To Be Published
|
|
7MWA
 
 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
4HFV
 
 | | Crystal structure of lpg1851 protein from Legionella pneumophila (putative T4SS effector) | | Descriptor: | CITRIC ACID, SUCCINIC ACID, Uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-11-07 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
5VKW
 
 | | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans | | Descriptor: | Adenylosuccinate lyase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans
To Be Published
|
|
5WP0
 
 | | Crystal structure of NAD synthetase NadE from Vibrio fischeri | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Stogios, P.J, Evdokimova, E, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of NAD synthetase NadE from Vibrio fischeri
To Be Published
|
|
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
5HNM
 
 | | Crystal structure of vancomycin resistance D,D-pentapeptidase VanY E175A mutant from VanB-type resistance cassette in complex with Zn(II) | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, SULFATE ION, ZINC ION | | Authors: | Stogios, P.J, Chun, J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
5UM7
 
 | | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii | | Descriptor: | ACETATE ION, Thioredoxin signature protein | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-26 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii
To Be Published
|
|
5V01
 
 | | Crystal structure of the competence damage-inducible protein A (ComA) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | CHLORIDE ION, Competence damage-inducible protein A, SODIUM ION | | Authors: | Borek, D, Wawrzak, Z, Grimshaw, S, Sandoval, J, Evdokimova, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the competence damage-inducible protein A (ComA) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
2I22
 
 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | Authors: | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2W
 
 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
6DM3
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | Descriptor: | RavO | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
7RCA
 
 | | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae | | Descriptor: | CHLORIDE ION, Chorismate synthase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae
To Be Published
|
|
7RGN
 
 | | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris
To Be Published
|
|
7REU
 
 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
7RGE
 
 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | Descriptor: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
7RJ1
 
 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
7RLL
 
 | | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate | | Descriptor: | Arf3p, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stogios, P.J, Michalska, K, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-25 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate
To Be Published
|
|
