8PFP
 
 | |
8PFL
 
 | | Crystal structure of WRN helicase domain in complex with 3 | | Descriptor: | 2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-7-oxidanylidene-6-[4-(3-oxidanylpyridin-2-yl)carbonylpiperazin-1-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]-~{N}-[2-methyl-4-(trifluoromethyl)phenyl]ethanamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
8PFO
 
 | | Crystal structure of WRN helicase domain in complex with HRO761 | | Descriptor: | Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION, ~{N}-[2-chloranyl-4-(trifluoromethyl)phenyl]-2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-6-[4-(6-methyl-5-oxidanyl-pyrimidin-4-yl)carbonylpiperazin-1-yl]-7-oxidanylidene-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]ethanamide | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
4YN2
 
 | | THE ATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, FUSOLIN, ZINC ION, ... | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YN1
 
 | | THE ATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, Fusolin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2R1R
 
 | |
6YHF
 
 | | Solution NMR Structure of APP TMD | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHO
 
 | | Solution NMR Structure of APP G38P mutant TM | | Descriptor: | Amyloid-beta precursor protein G38P mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHP
 
 | | Solution NMR Structure of APP V44M mutant TMD | | Descriptor: | Amyloid-beta precursor protein V44M mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHX
 
 | | Solution NMR Structure of APP I45T mutant TMD | | Descriptor: | Amyloid-beta precursor protein I45T mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHI
 
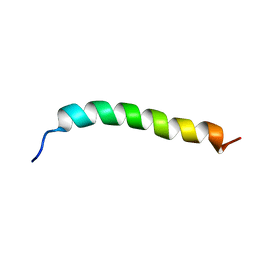 | | Solution NMR Structure of APP G38L mutant TMD | | Descriptor: | Amyloid-beta precursor protein G38L mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
2BXJ
 
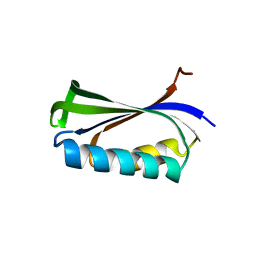 | | Double Mutant of the Ribosomal Protein S6 | | Descriptor: | 30S RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antagonism, Non-Native Interactions and Non-Two-State Folding in S6 Revealed by Double-Mutant Cycle Analysis.
Protein Eng.Des.Sel., 18, 2005
|
|
6Z9I
 
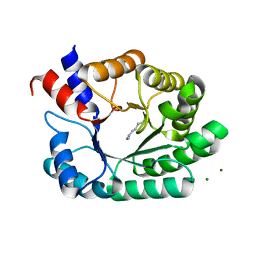 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - N21K mutant complex with reaction products | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6Z9J
 
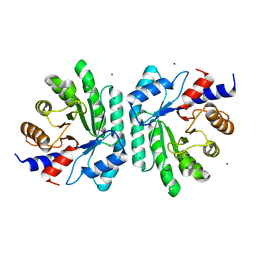 | |
6Z9H
 
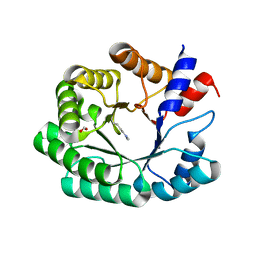 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - C47V/G204A/S239D mutant | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, FORMIC ACID, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
7ZAK
 
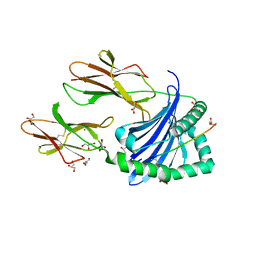 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7PZR
 
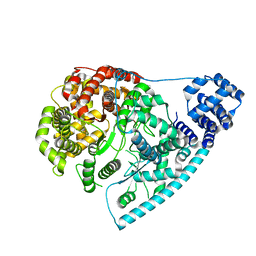 | | Cryo-EM structure of POLRMT in free form. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
5WDT
 
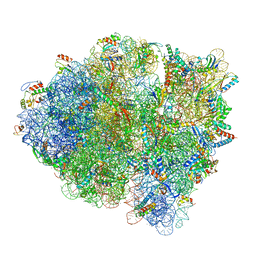 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WE6
 
 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
7PZP
 
 | |
5WE4
 
 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLK
 
 | |
7QLL
 
 | |
