5GN9
 
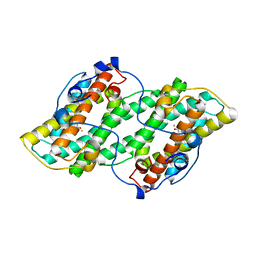 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN5
 
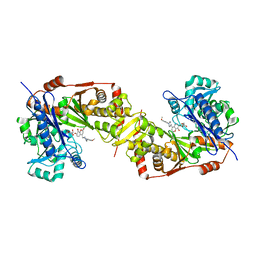 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
1LUE
 
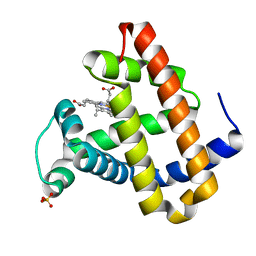 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
1O16
 
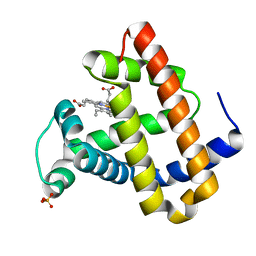 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68S/D122N MUTANT (MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-10-25 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6KYP
 
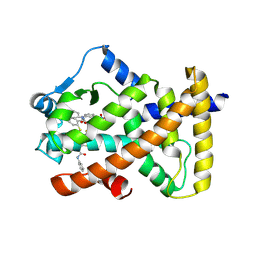 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
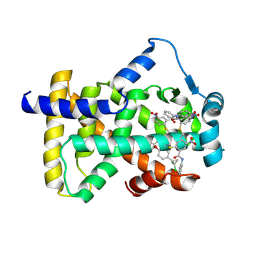 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L1B
 
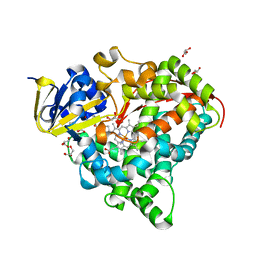 | | Crystal Structure of P450BM3 with N-(3-cyclopentylpropanoyl)-L-pipecolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-(3-cyclopentylpropanoyl)piperidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6L36
 
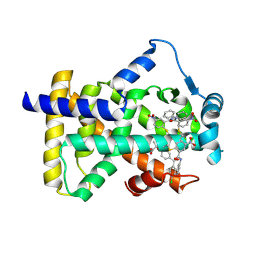 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5SIC
 
 | |
8Y11
 
 | |
8Y4J
 
 | |
8XWK
 
 | |
8Y4B
 
 | |
8Y46
 
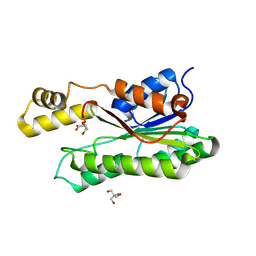 | | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase bound to L-KDF or L-2,4-DKDF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, L-2,4-diketo-3-deoxyfuconate, ... | | Authors: | Akagashi, M, Watanabe, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase reveals a unique binding mode as a alpha-furanosyl hemiketal of substrates.
Sci Rep, 14, 2024
|
|
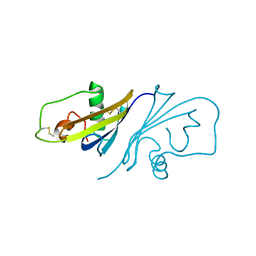
3SSI
 
 | |
2SIC
 
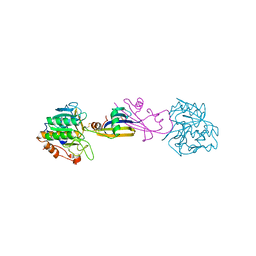 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
1MPG
 
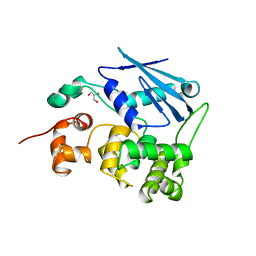 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
3NG5
 
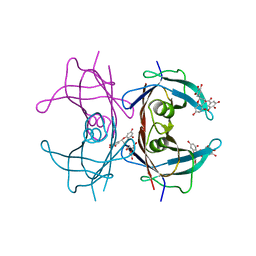 | | Crystal Structure of V30M transthyretin complexed with (-)-epigallocatechin gallate (EGCG) | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, GLYCEROL, Transthyretin | | Authors: | Miyata, M, Nakamura, T, Ikemizu, S, Chirifu, M, Yamagata, Y, Kai, H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of green tea polyphenol(-)-epigallocatechin gallate (EGCG)-transthyretin complex reveals novel binding site distinct from thyroxine binding site
Biochemistry, 2010
|
|
6XOX
 
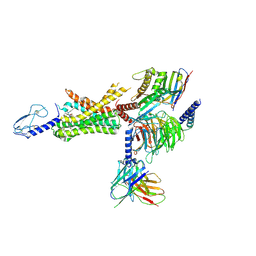 | | cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970 | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, Alpha subunit of Gs with N-terminus swapped with equivalent residues in Gi,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4YD9
 
 | | Crystal structure of squid hemocyanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CU2-O2 CLUSTER, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuno, A, Gai, Z, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-21 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 3.8-MDa Respiratory Supermolecule Hemocyanin at 3.0 angstrom Resolution
Structure, 23, 2015
|
|
