8SAI
 
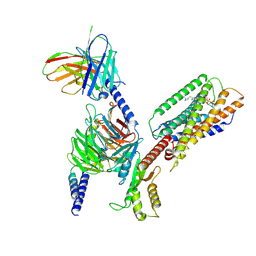 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HK5
 
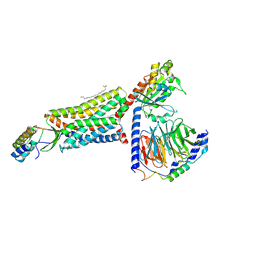 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
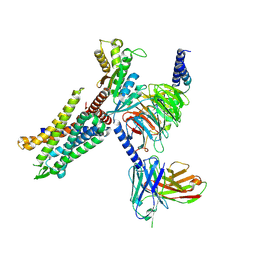 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK2
 
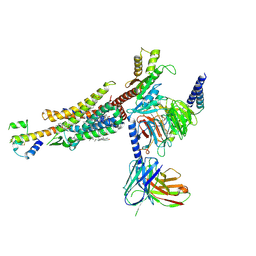 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
5TWM
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
7BST
 
 | | EcoR124I-Ocr in the Intermediate State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTR
 
 | | EcoR124I-ArdA in the Restriction-Alleviation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTQ
 
 | | EcoR124I-DNA in the Restriction-Alleviation State | | Descriptor: | DNA (64-MER), Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTO
 
 | | EcoR124I-ArdA in the Translocation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTP
 
 | | EcoR124I-Ocr in Restriction-Alleviation State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
3VOD
 
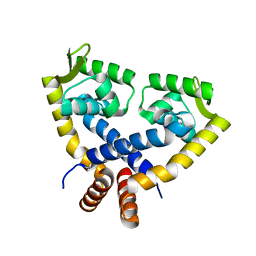 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
5VQF
 
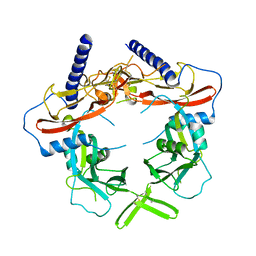 | | Crystal Structure of pro-TGF-beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
5H3R
 
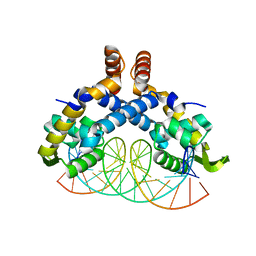 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | Authors: | Zhu, R, Lou, H, Hao, Z. | | Deposit date: | 2016-10-26 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
3ML8
 
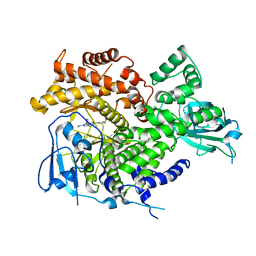 | |
4CMO
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 2-((1R)-1-((3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl) oxy)ethyl)-4-fluoro-N-methylbenzamide | | Descriptor: | 2-[(1R)-1-{[3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl]oxy}ethyl]-4-fluoro-N-methylbenzamide, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CTC
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17- dihydro-1H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecin-15(10H)-one | | Descriptor: | (10R)-7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17-dihydro-1H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
3ML9
 
 | |
4CTB
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20- tetrahydro-7,11-(azeno)pyrido(2',1':2,3)imidazo(4,5-h)(2,5,11) benzoxadiazacyclotetradecine-14-carbonitrile | | Descriptor: | (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20-tetrahydro-11,7-(azeno)pyrido[2',1':2,3]imidazo[4,5-h][2,5,11]benzoxadiazacyclotetradecine-14-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CLJ
 
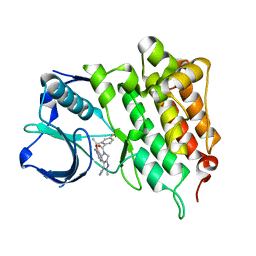 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CLI
 
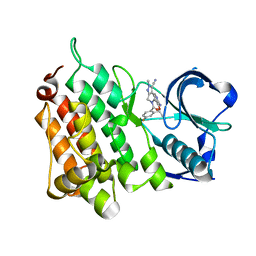 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with PF- 06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16, 17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CNH
 
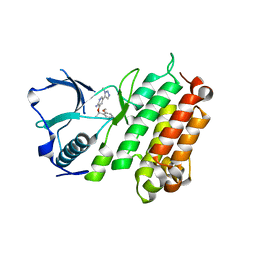 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy)-5-(1-methyl-1H- 1,2,3-triazol-5-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy]-5-(1-methyl-1H-1,2,3-triazol-5-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CCU
 
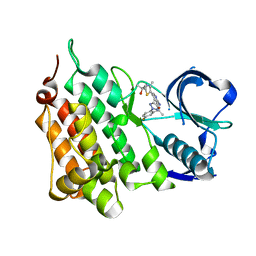 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy) pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol | | Descriptor: | 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
