8HCZ
 
 | |
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
2HQV
 
 | | X-ray Crystal Structure of Protein AGR_C_4470 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR92. | | Descriptor: | AGR_C_4470p | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Zhao, L, Cunningham, K, Ma, L.C, Fang, Y, Xiao, R, Acton, T, Montelione, T.G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AGR_C_4470p from Agrobacterium tumefaciens.
Protein Sci., 16, 2007
|
|
8X8L
 
 | | Cryo-EM structure of the cortistatin 17-bound Somatostatin receptor 5-Gi protein complex | | Descriptor: | Cortistatin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, H.E, You, C, Zhao, L, Li, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for activation of somatostatin receptor 5 by cyclic neuropeptide agonists.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8N
 
 | | Cryo-EM structure of the octreotide-bound Somatostatin receptor 5-Gi protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha, ... | | Authors: | Xu, H.E, You, C, Zhao, L, Li, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for activation of somatostatin receptor 5 by cyclic neuropeptide agonists.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2HD3
 
 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4HUF
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*TP*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUE
 
 | | Structure of 5-chlorouracil modified G:U base pair | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(UCL)P*GP*CP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HYI
 
 | | X-RAY Crystal structure of compound 40 bound to human chk1 kinase domain | | Descriptor: | 2-(1H-indazol-1-yl)-N-[2-(piperazin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure-based design and optimization of 2-aminothiazole-4-carboxamide as a new class of CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
6NKH
 
 | |
6NKK
 
 | | Structure of PhqE Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and premalbrancheamide | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NKI
 
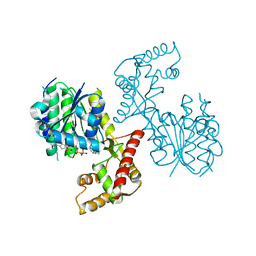 | | Structure of PhqB Reductase Domain from Penicillium fellutanum | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NRPS | | Authors: | Dan, Q, Newmister, S.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NKM
 
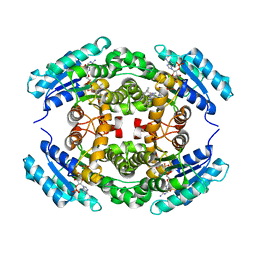 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | Descriptor: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
4HYH
 
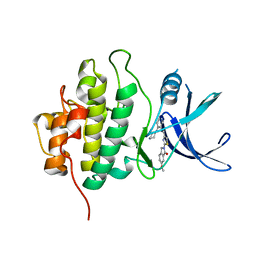 | | X-RAY Crystal structure of compound 39 bound to human chk1 kinase domain | | Descriptor: | 2-(6-methoxy-1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-[4-(piperazin-1-yl)pyridin-3-yl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design and optimization of 2-aminothiazole-4-carboxamide as a new class of CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7FEJ
 
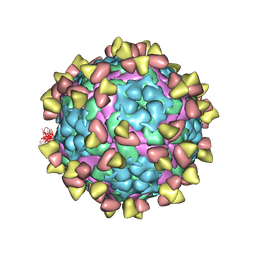 | | Complex of FMDV A/AF/72 and bovine neutralizing scFv antibody R55 | | Descriptor: | A/AF/72 VP1, A/AF/72 VP2, A/AF/72 VP3, ... | | Authors: | He, Y, Li, K, Lou, Z. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of Foot-and-Mouth Disease Virus with Bovine Neutralizing Antibodies Reveal the Determinant of Intraserotype Cross-Neutralization.
J.Virol., 95, 2021
|
|
7FEI
 
 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody R55 | | Descriptor: | Capsid protein VP0, IG HEAVY CHAIN VARIABLE REGION, IG LAMDA CHAIN VARIABLE REGION | | Authors: | He, Y, Li, K, Lou, Z. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of Foot-and-Mouth Disease Virus with Bovine Neutralizing Antibodies Reveal the Determinant of Intraserotype Cross-Neutralization.
J.Virol., 95, 2021
|
|
5YFG
 
 | | SOLUTION STRUCTURE OF HUMAN MOG1 | | Descriptor: | Ran guanine nucleotide release factor | | Authors: | Hu, Q, Liu, Y, Bao, X, Liu, H. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation
J Mol Cell Biol, 10, 2018
|
|
5WDJ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH COMPOUND-6 AKA 7-(BENZYLOXY)-1H-[1,2, 3]TRIAZOLO[4,5-D]PYRIMIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(benzyloxy)-1H-[1,2,3]triazolo[4,5-d]pyrimidin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2017-07-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyrimidines identified as reversible myeloperoxidase inhibitors.
Medchemcomm, 8, 2017
|
|
5W2A
 
 | |
7E2P
 
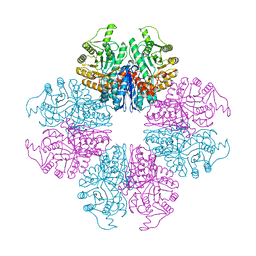 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2Q
 
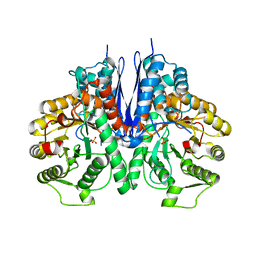 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7TVA
 
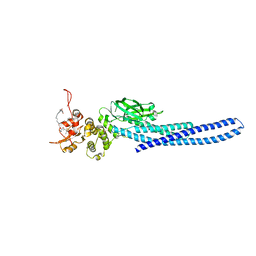 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
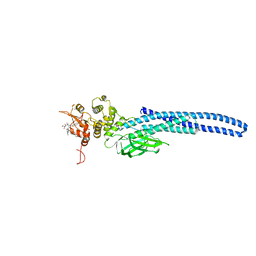 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
