8GMP
 
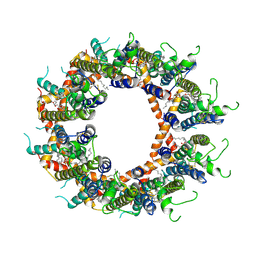 | | Cryo-EM structure of octameric human CALHM1 with a I109W point mutation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calcium homeostasis modulator protein 1 | | Authors: | Syrjanen, J.L, Furukawa, H. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of human CALHM1 reveals key locations for channel regulation and blockade by ruthenium red.
Nat Commun, 14, 2023
|
|
8GI9
 
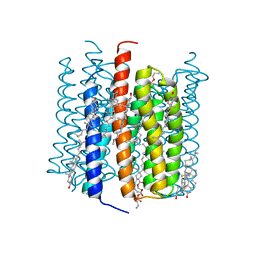 | | Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Cation Channelrhodopsin, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8GA1
 
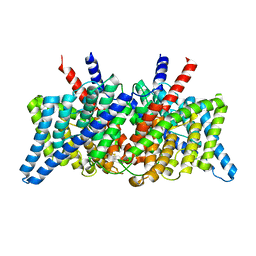 | | CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Swap | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GA3
 
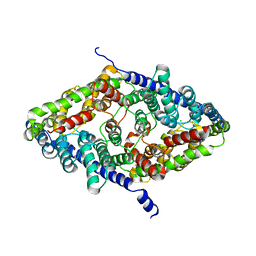 | | CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Turn | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GAH
 
 | | CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Twist | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GA0
 
 | | CLC-ec1 E202Y at pH 4.5 100mM Cl Turn | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GA5
 
 | | CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Intermediate | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQE
 
 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
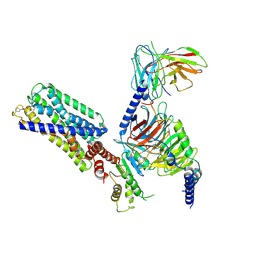 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
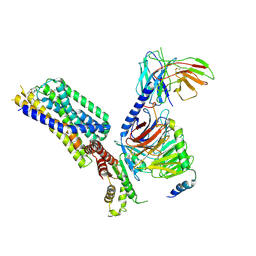 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8I9O
 
 | |
8IMA
 
 | | Filament structure of GAC with phosphate | | Descriptor: | Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Guo, C.J, Liu, J.L. | | Deposit date: | 2023-03-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for activation and filamentation of glutaminase.
Cell Res., 34, 2024
|
|
8IMB
 
 | | Filament interface structure of GAC with phosphate | | Descriptor: | Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Guo, C.J, Liu, J.L. | | Deposit date: | 2023-03-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for activation and filamentation of glutaminase.
Cell Res., 34, 2024
|
|
8J0G
 
 | |
8J0E
 
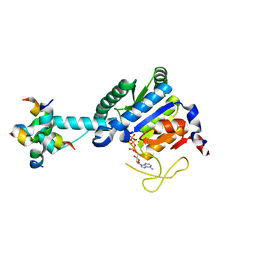 | | GK monomer complexes with catalytic intermediate | | Descriptor: | (2~{R})-5-[[[[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2-azanyl-5-oxidanylidene-pentanoic acid, Delta-1-pyrroline-5-carboxylate synthase B, MAGNESIUM ION | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | GK monomer complexes with catalytic intermediate
To Be Published
|
|
8J27
 
 | | GK monomer complexes with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GK monomer complexes with ADP
To Be Published
|
|
8J28
 
 | | GK monomer complexes with G5P and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GK monomer complexes with G5P and ADP
To Be Published
|
|
8J0F
 
 | |
1T9P
 
 | | Crystal Structure of V44A, G45P Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1T0W
 
 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1T9Q
 
 | | Crystal Structure of V44L Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
6YCH
 
 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
