3VZM
 
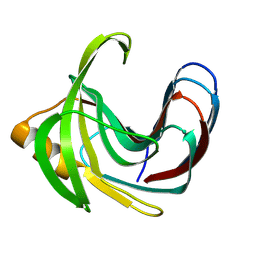 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZK
 
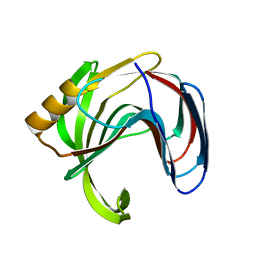 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZO
 
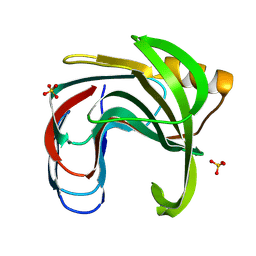 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3UHI
 
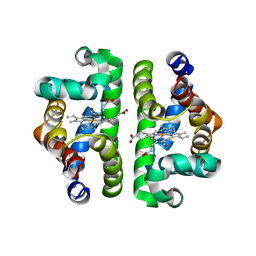 | | HBI (K96R) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHW
 
 | | HBI (N79A) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UGZ
 
 | | HBI (L36A) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH7
 
 | | HBI (T72G) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH3
 
 | | HBI (L36V) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHB
 
 | | HBI (R104K) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHZ
 
 | | HBI (T72A) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5Y8R
 
 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6ARK
 
 | | Crystal Structure of compound 10 covalently bound to K-Ras G12C | | Descriptor: | (3R)-N-(6-bromonaphthalen-2-yl)-3-hydroxy-1-propanoyl-L-prolinamide, GLYCEROL, GTPase KRas, ... | | Authors: | Nnadi, C.I, Jenkins, M.L, Gentile, D.R, Bateman, L.A, Zaidman, D, Balius, T.E, Nomura, D.K, Burke, J.E, Shokat, K.M, London, N. | | Deposit date: | 2017-08-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange.
J Chem Inf Model, 58, 2018
|
|
5Z6V
 
 | |
6BB4
 
 | |
6APX
 
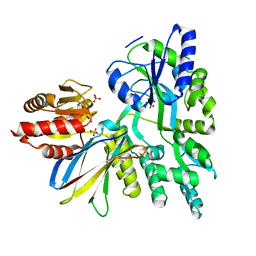 | | Crystal structure of human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the monobody YSX1 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Dual specificity protein phosphatase 1, Monobody YSX1, ... | | Authors: | Gumpena, R, Lountos, G.T, Sreejith, R.K, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Protein Sci., 27, 2018
|
|
6B3X
 
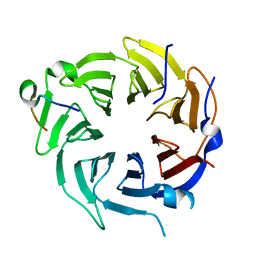 | | Crystal structure of CstF-50 in complex with CstF-77 | | Descriptor: | Cleavage stimulation factor subunit 1, Cleavage stimulation factor subunit 3 | | Authors: | Yang, W, Hsu, P, Yang, F, Song, J.E, Varani, G. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Nucleic Acids Res., 46, 2018
|
|
6BN8
 
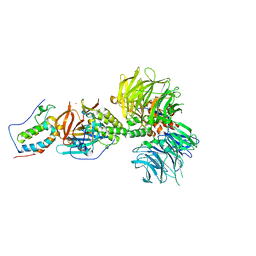 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
2XGA
 
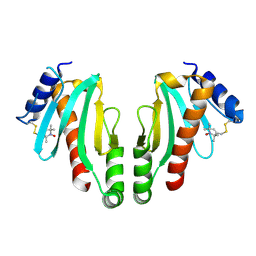 | | MTSL spin-labelled Shigella Flexneri Spa15 | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAK | | Authors: | Lillington, J.E.D, Johnson, S, Lea, S.M. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
J.Mol.Biol., 405, 2011
|
|
3DA2
 
 | | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 13, N-(4-chlorobenzyl)-N-methylbenzene-1,4-disulfonamide, ... | | Authors: | Pilka, E.S, Picaud, S.S, Yue, W.W, King, O.N.F, Bray, J.E, Filippakopoulos, P, Roos, A.K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor.
To be Published
|
|
7PP7
 
 | | Thunberia alata 16:0-ACP desaturase | | Descriptor: | Acyl-[acyl-carrier-protein] 6-desaturase, FE (III) ION | | Authors: | Guy, J.E, Whittle, E, Cai, Y, Chai, J, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2021-09-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselectivity mechanism of the Thunbergia alata Delta 6-16:0-acyl carrier protein desaturase.
Plant Physiol., 188, 2022
|
|
3DS9
 
 | |
3DDX
 
 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
