5BYA
 
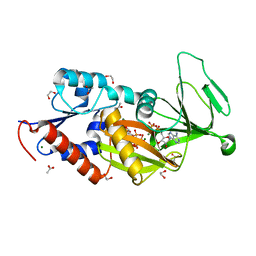 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthetic tools for studying the chemical biology of InsP8.
Chem.Commun.(Camb.), 51, 2015
|
|
5B6O
 
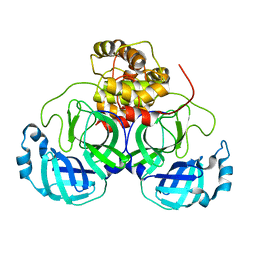 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8JHO
 
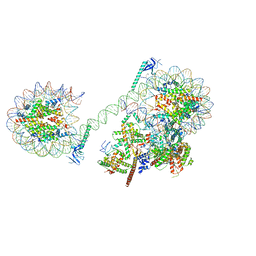 | |
2QLK
 
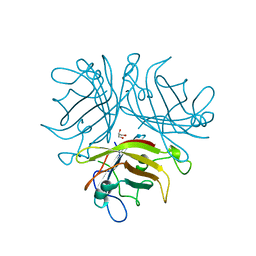 | | Adenovirus AD35 fibre head | | Descriptor: | Fiber, GLYCEROL | | Authors: | Liaw, Y.-C, Amiraslanov, I, Wang, H, Lieber, A. | | Deposit date: | 2007-07-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob
J.Virol., 81, 2007
|
|
6N5C
 
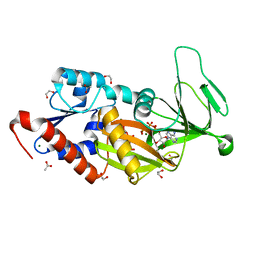 | | Crystal structure of the catalytic domain of PPIP5K2 in complex with AMPPNP and 5-PCF2Am-InsP5 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B, Riley, A, Potter, B. | | Deposit date: | 2018-11-21 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of an alpha-phosphono-alpha , alpha-difluoroacetamide analogue of the diphosphoinositol pentakisphosphate 5-InsP7.
Medchemcomm, 10, 2019
|
|
4Q4C
 
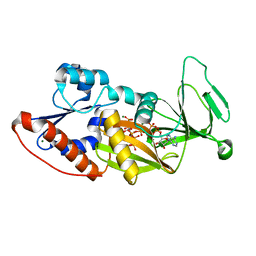 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and synthetic 1,5-(PP)2-IP4 (1,5-IP8) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4Q4D
 
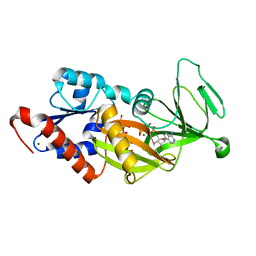 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMP-PNP and synthetic 3,5-(PP)2-IP4 (3,5-IP8) | | Descriptor: | (1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2DUC
 
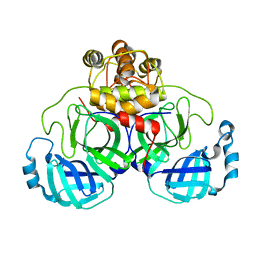 | | Crystal structure of SARS coronavirus main proteinase(3CLPRO) | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Wang, H, Kim, Y.T, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-21 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4HN2
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and a substrate analog 5PA-IP5 | | Descriptor: | (2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}ethyl)phosphonic acid, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H. | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First synthetic analogues of diphosphoinositol polyphosphates: interaction with PP-InsP(5) kinase.
Chem.Commun.(Camb.), 48, 2012
|
|
7YRN
 
 | |
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
6AQ4
 
 | | CRYSTAL STRUCTURE OF PROTEIN CiTE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND CITRAMALYL-COA | | Descriptor: | CHLORIDE ION, Citramalyl-CoA, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ARB
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, PYRUVATE AND COENZYME A | | Descriptor: | COENZYME A, Citrate lyase subunit beta-like protein, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AS5
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, ACETOACETATE AND COENZYME A | | Descriptor: | ACETOACETIC ACID, COENZYME A, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
8IRW
 
 | | Structure of nanobody Nb9 against parathion | | Descriptor: | nanobody Nb9 against parathion | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2023-03-19 | | Release date: | 2024-03-20 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Evolution of Antiparathion Nanobody with Enhanced Sensitivity and Specificity Based on Structural Analysis.
J.Agric.Food Chem., 71, 2023
|
|
8G9E
 
 | |
8ZBL
 
 | | Glycosidated glycyrrhetinic acid derivative as a soluble epoxide hydrolase inhibitor | | Descriptor: | 1-[[(2~{S},4~{a}~{S},6~{a}~{R},6~{a}~{S},6~{b}~{R},8~{a}~{R},10~{S},12~{a}~{S},14~{b}~{S})-10-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,4~{a},6~{a},6~{b},9,9,12~{a}-heptamethyl-13-oxidanylidene-3,4,5,6,6~{a},7,8,8~{a},10,11,12,14~{b}-dodecahydro-1~{H}-picen-2-yl]methyl]-3-(oxan-4-ylmethyl)urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Feng, Y, Ge, Z.H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of glycosidated glycyrrhetinic acid derivatives: Natural product-based soluble epoxide hydrolase inhibitors.
Eur.J.Med.Chem., 280, 2024
|
|
1V3R
 
 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
8CEO
 
 | |
8CEN
 
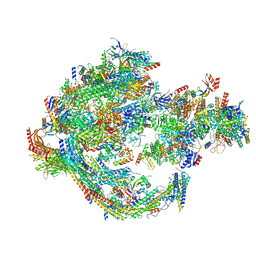 | | Yeast RNA polymerase II transcription pre-initiation complex with core Mediator | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, H, Schilbach, S, Cramer, P. | | Deposit date: | 2023-02-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Yeast PIC-Mediator structure with RNA polymerase II C-terminal domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
