2F4M
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G9F
 
 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6DLO
 
 | |
4LNY
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
6A2H
 
 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(P*AP*AP*TP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*T)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
4LT9
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
6A2I
 
 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*CP*GP*TP*AP*GP*CP*TP*AP*AP*TP*TP*AP*GP*CP*TP*AP*CP*G)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
5Y9Q
 
 | |
1OS5
 
 | | Crystal structure of HCV NS5B RNA polymerase complexed with a novel non-competitive inhibitor. | | Descriptor: | 3-(4-AMINO-2-TERT-BUTYL-5-METHYL-PHENYLSULFANYL)-6-CYCLOPENTYL-4-HYDROXY-6-[2-(4-HYDROXY-PHENYL)-ETHYL]-5,6-DIHYDRO-PYRAN-2-ONE, Hepatitis C virus NS5B RNA polymerase | | Authors: | Love, R.A, Parge, H.E, Yu, X, Hickey, M.J, Diehl, W, Gao, J, Wriggers, H, Ekker, A, Wang, L, Thomson, J.A, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2003-03-18 | | Release date: | 2004-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme.
J.Virol., 77, 2003
|
|
2YNI
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK952 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNF
 
 | | HIV-1 Reverse Transcriptase Y188L mutant in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNG
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
3TC6
 
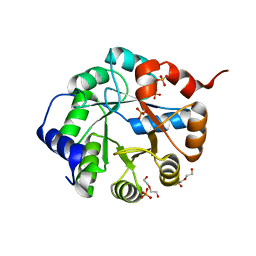 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
8WY4
 
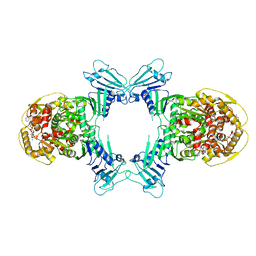 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
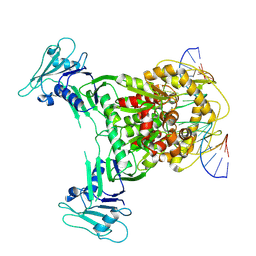 | | Structure of DNA-bound GajA dimer (focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | Structure of ATP/Mg2+ bound Gabija GajA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
2ODA
 
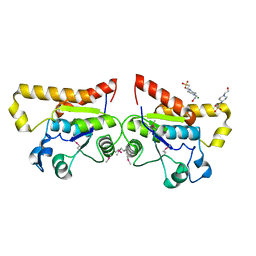 | | Crystal Structure of PSPTO_2114 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | Authors: | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | Deposit date: | 2006-12-22 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
8X5N
 
 | | Structure of ATP/Mg2+ bound Gabija GajA-GajB 4:4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6DLP
 
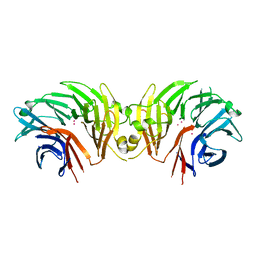 | | Crystal structure of LRRK2 WD40 domain dimer | | Descriptor: | Leucine-rich repeat serine/threonine-protein kinase 2, PLATINUM (II) ION | | Authors: | Zhang, P, Ru, H, Wang, L, Wu, H. | | Deposit date: | 2018-06-02 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the WD40 domain dimer of LRRK2.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8V6U
 
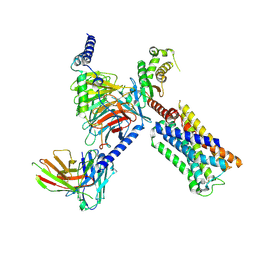 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
7VFB
 
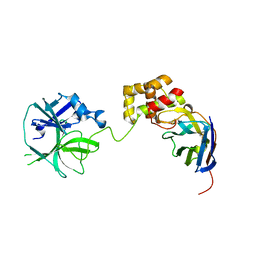 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFA
 
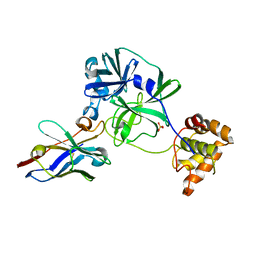 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
